iGEM REPORT: Engineering a ‘BioBalloon’ for Mid-Atmospheric Sensing: Synthetic Biological Applications of Latex, Melanin, Chlamydomonas reinhardtii, Nucleic Acid Aptamers, and Chromogenic Proteins
Note: This iGEM Report was submitted to the PLOS iGEM Realtime Peer Review Jamboree, and has not undergone formal peer review by any of the PLOS journals. We welcome your comments on this work.
Engineering a ‘BioBalloon’ for Mid-Atmospheric Sensing: Synthetic Biological Applications of Latex, Melanin, Chlamydomonas reinhardtii, Nucleic Acid Aptamers, and Chromogenic Proteins
Michael J. Becich (1) *, Gordon L. Sun (1), Cynthia A. Hale-Phillips (2), Taylor N. Sihavong (1), Theresa C. Sievert (1), Hieu-Thao Anna Le (1), Amy S. Weissenbach (1), Charles R. Gleason (2), Elias G. Robinson (2), Eric Y. Liu (2), Trevor! Kalkus (3), Kara J. Helmke Rogers (1), Lynn J. Rothschild (4) *
- Department of Bioengineering, Stanford University, Stanford, CA USA 94305
- Brown University, Providence, RI USA 02912
- Universities Space Research Association at NASA Ames Research Center, Moffett Field, CA USA 94035
- NASA Ames Research Center, Moffett Field, CA USA 94035
*Corresponding author: Michael Becich (mbecich@stanford.edu) [student], Dr. Lynn Rothschild (lynn.j.rothschild@nasa.gov)
Author Contributions
Conceived and designed the experiments: GLS CAHP CRG MJB LJR ASW TK
Performed the experiments: CAHP TCS TNS EGR GLS CRG MJB ASW HTAL EYL TK
Analyzed the data: GLS CAHP TNS TCS CRG MJB EGR ASW KJHR HTAL
Wrote the paper: MJB TNS GLS CAHP TCS EGR LJR ASW HTAL
Reviewed the paper: MJB TNS HTAL EGR GLS CAHP TCS CRG LJR ASW KJHR TK EYL
Abstract
Atmospheric research – whether on Earth or beyond – broadens our understanding of atmospheric processes, including those critical for formation and sustenance of life on a planet’s surface. Using synthetic biology, we have prototyped a novel, inexpensive, and lightweight atmospheric exploration and monitoring platform–a biomaterials-based balloon (BioBalloon). Our approach focused on four key areas: (1) membrane material production, (2) membrane atmospheric protection, (3) gas production, and (4) biosensor design. For membrane material construction, E. coli was metabolically engineered to convert glucose to a latex precursor, as synthetic rubber is the material choice for high-altitude balloons on Earth. To mitigate risk of membrane damage by solar radiation, we engineered E. coli to overexpress melanin, a UV-absorbing pigment, and designed new linker proteins in order to integrate melanin into the membrane superstructure. We demonstrated controlled production of hydrogen and oxygen gas by the green alga Chlamydomonas reinhardtii. Finally, to increase sensing area and decrease payload mass, we designed and tested sensors to both be carried by a traditional payload, and be attached to the membrane. Biosensor design focused on developing a cassette of thermosensitive chromoproteins for temperature sensing and an aptamer-based fluorophore-quenching system to detect biosignatures like ATP. When assembled, these components can be used to construct an entirely biosynthetic BioBalloon.
Financial Disclosure
The Stanford-Brown iGEM team is incredibly grateful for financial support from the NASA Space Technology Mission Directorate Early Stage Innovation program, Stanford VPUE Grant for Undergraduate Research to the Stanford Bioengineering Department, the Brown University UTRA, and the Rhode Island Space Grant Consortium for continued support for the Stanford-Brown team. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing Interests
The authors have declared that no competing interests exist.
Data Availability
In alignment with the PLOS data sharing policy, all data, protocols, figures, videos, and documentation are fully available without restriction: http://2016.igem.org/Team:Stanford-Brown.
Introduction
Terrestrial and extraterrestrial atmospheres help us understand climate dynamics, atmospheric chemistry evolution, wind patterns and weather patterns. These in turn may help us understand the habitability of a planet, whether on the surface or in the air itself. For atmospheric exploration, high-altitude weather balloons are the most commonly used research vehicle for tracking weather and wind patterns, and for monitoring atmospheric composition. However, large-scale sampling of atmospheric properties is not currently economically feasible, and contemporary weather balloons have limited payload capacity. Today, planetary scientists hope to conduct atmospheric research on other planets, but they lack adequate tools. Planetary scientists are particularly interested in further exploring the effects of greenhouse gases on the volatile atmosphere of Venus, the abundance of carbon dioxide on Mars and the implications of its thin atmosphere for sustaining life, and Titan’s resemblance to Earth in nitrogen enveloping hydrogen dioxide. However, current planetary research depends upon orbiters, which monitor planets at a distance, and rovers, which monitor the planet surface. Planetary scientists lack a research tool for atmospheric research. Such a tool would need to both traverse extensive vertical and horizontal distances and collect relevant atmospheric data.
Here, we propose a “BioBalloon,” or a biologically produced balloon, which would harness and expand upon the capabilities of a balloon. The balloon could be made in situ using engineered bacteria which would in theory allow unlimited production on site. It could contain temperature and small-molecule sensors, increasing sensing surface area and decreasing payload weight. To begin to prototype such a balloon, our team developed a biomaterial-based balloon by engineering synthetic organisms for sensor and balloon membrane production. To make the balloon itself, E. coli was metabolically engineered to produce elastic polymers for incorporation into the membrane structure. Additional functionality was achieved by engineering biological temperature and small molecule biosensors, which could be imbedded in the balloon membrane via biotinylation. When combined, these components can be constructed into a balloon made entirely out of biological products.
Membrane
While most naturally occurring membrane vacuoles are optimized for storing fluids, few are used to store gas. For a membraneous compartment to store gas, it needs to resist both leakage and variation in internal pressure. Furthermore, for atmospheric exploration purposes, the membrane must be durable enough to withstand debris and other potential damages to the structure. Therefore, research was done on both durable and resilient fibers that could be incorporated into the balloon membrane.
The durability of p-aramid fibers can be utilized to meet the specific needs of inflatable modules and habitats. The sturdiness and low density of the material allows for construction of a lightweight structural reinforcement for a balloon membrane. Furthermore, p-aramid’s resistance to chemical, thermal, and physical wear make it ideal for minimizing material corrosion. Currently p-aramid fiber analogues are used for strengthening outer coverings on existing space modules to protect them against exposure in space [1]. To engineer a membrane with these desirable properties, we identified and isolated E. coli genes in the Shikimate pathway relevant for para-aminobenzoic acid [2], a p-aramid monomer analogue.
Resilience in membrane material was explored by investigating potential biological and synthetic membranes. The natural solution to the high pressures and repeated expansion and contraction can be found in proteins that make up the connective tissue within the human arterial wall. The combination of the two predominant proteins in this tissue, elastin and collagen, can provide the membrane both properties of elasticity and strength [3]. Aiming to create a recombinant membrane that shared the biological structure’s behavior, the team designed and synthesized elastin and collagen monomers in E. coli.
Due to its large stretch ratio and high resilience to repeated stress, latex is a strong synthetic candidate material for membrane construction. Used extensively as both a major structural and dampening component in the construction of space exploration vehicles, latex is a critical building material for furthering space exploration [4]. Additionally, latex’s high flexibility and tensile strength make it an appealing material for construction of flexible structures that can withstand variable mechanical stresses. However, use of rubber materials in space is currently limited by cost of bulk material transport [5]—thereby illuminating the need for a sustainable, scalable, and transportable method of latex production.
Latex is commonly produced either through chemical synthesis of its primary constituent, polyisoprene chains, or natural harvesting from rubber tree plantations. However, chemical synthesis and rubber farming have been threatened by production shortfalls due to rising petroleum prices, disease, and deforestation. These methods are also limited in production overhead, sustainability, scale, and turnaround, highlighting a need for an alternative method for latex production [6, 7]. To address this need, we engineered the Hevea brasiliensis polyisoprene synthesis pathway into E. coli for production of latex polymers. These polymers could then be constructed into membranes for building a BioBalloon, and the ability to grow rubber-producing cultures from a single cell could provide a sustainable and renewable source of a complex building material in space.
To generate a composite membrane durable enough to store hydrogen (or any gas capable of generating lift) in variable atmospheric conditions, p-aramid fibers were incorporated into membrane design to increase membrane tensile strength, as they exhibit a low density yet high tensile strength ideal for fortifying elastic membranes. Furthermore, for resilience properties, a membrane of elastin and collagen was synthesized in vivo to harness their combined dynamic properties. Latex was synthesized bacterially as an additional membrane candidate.
Membrane protection
Increased exposure to UV radiation at high altitudes can compromise balloon structure; extended UV exposure can disrupt the molecular interactions within thin membranes, creating micropores that decrease the material’s structural integrity and impermeability. On Earth, the main source of concern for radiation damage is ultraviolet radiation (UVR). These wavelengths are between 290 and 400 nm when they hit the Earth’s surface, and do more damage the shorter they get. To install radiation resistance in our biomembranes, the team investigated the absorptive properties of melanin. Melanin is the primary source of naturally-occurring UV protection in human skin. In response to human skin damage through UV exposure, keratinocytes release cytokines to stimulate melanocytes’ production of melanin, which functions as a UV absorbent and generates the phenotypic response of darker skin [8]. Melanin when used as a sunscreen has been shown to absorb between 50-75% of UVR [9]. We decided to focus on creating a method of UV protection for our balloon that utilizes melanin incorporated via a binding agent directly into our membranes, and designed a novel binding mechanism to incorporate the pigment directly into material.
Biogas production
To generate lift, a balloon must contain a buoyant lifting force in excess of gravitational pull. An important consideration in balloon implementation as an atmospheric sensing vehicle is the mechanism for lift, which for a balloon would be a low density gas. On a planet like Mars, surface pressure is only 0.6% of Earth’s which means heavier gases may not provide as much lift [10]. Hydrogen gas is a lightweight alternative, and can be produced by the green alga Chlamydomonas reinhardtii. Albeit slower than electrohydrolysis, the slower rate of gas production allows for precision control of balloon filling, where excess gas can be easily bled. C. reinhardtii also produces both oxygen and hydrogen gas, which can be modulated to yield different gas compositions for use in a variety of planetary atmospheres [11].
Sensing
To add functionality to the balloon, a cassette of chromoproteins was characterised for use as part of a bio-thermometer. Chromogenic proteins (chromoproteins) are colored proteins that have been extracted from a variety of organisms such as the sea anemone, Actinia equine [12], or have been synthetically produced, as was the case for the chromoproteins from ATUM (Newark, CA). Like all proteins, chromogenic proteins are sensitive to heat; however, their sensitivity to heat is coupled with a visible color loss or change. The basic structure of a chromogenic protein consists of a large beta barrel and a chromophore which is supported by the beta barrel. The chromophore interacts with light and gives the protein its distinctive color. Additionally, the shape of the beta barrel is supported by water molecules that interact with the side chains of the amino acids that the beta barrel comprises of [13]. As the research herein confirms, different chromoproteins respond to different temperatures by either losing their color or changing to a different color. In planetary exploration, monitoring temperature changes in the atmosphere is important for predicting weather patterns and atmospheric stability, which vary drastically with respect to altitude. Thus, a precise biological temperature sensor must be developed in order to address these concerns. The biological temperature sensor presented herein can be expressed in a small culture of E. coli and is easily maintained and replaced.
As additional functionality, an ideal BioBalloon would have small molecule sensing capabilities to monitor atmospheric composition, searching for biosignatures, toxins, minerals, and/or pathogens. A fluorophore-quencher (FQ) system was developed to sense ATP, a proxy for terrestrial life. The FQ system uses a single stranded DNA aptamer sensing domain and fluorescence as an expression platform to signal that the aptamer had bound its target [14]. After testing the sensor’s function while bound to a streptavidin plate, a pre-characterized cellulose cross-linker [15] was added and tested while was bound to a proxy cellulose “balloon membrane.”
Materials and Methods
Building a membrane with latex rubber
Plasmid assembly
Plasmids were assembled into the International Genetically Engineered Machine (iGEM) standard plasmid backbones pSB1C3 and pSB2K3. Gene inserts were synthesized by Integrated DNA Technologies (6024 Silver Creek Valley Road, San Jose, CA 95138 USA) as gBlocks, and assembled according to Gibson Assembly Protocol standards with NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs, Ipswich, MA.) Due to similarity in ORI between pSB1C3 and pSB2K3 however, pUC19 was used as a backbone in place of the latter for the double transformation.
DNA sequencing and analysis
Liquid cultures of NEB 5-alpha cells transformed with the two latex production plasmids were run through a Qiagen Miniprep Kit to isolate plasmid DNA constructs. Purified constructs were then sequenced for each of the four inserts, using ELIM premix sequencing standards. Samples were then sequenced by ELIM Biopharmaceuticals (Hayward, CA, USA), and chromatogram results were analyzed and aligned using Geneious (Biomatters, San Francisco, CA, USA).
Cell Culture
In order to produce latex, the materials needed are transformation-competent E. coli cells, LB growth media, aqueous MgSO4 or another magnesium (II) supplement, the two mentioned plasmids for transfection and/or any variation, glucose solution and any standard cell transformation kit. Bioreactors can be used to culture large volumes of cells to maximize polyisoprene yield. NEB 5-alpha and T7 Express Competent E. coli strains used in all transformation experiments were acquired from New England Biolabs (Ipswich, MA). Liquid cultures were grown in LB medium and incubated overnight at 37 °C at 200 RPM. IPTG was added after 18 hours of incubation to T7 cells transformed with the DXS synthase operon and the gene cassette containing both cis-prenyltransferases and SRPP induce expression. To induce latex production, IPTG (400 mM), glucose (1 M), and MgSO4 (400 mM) were added directly to 350 ml liquid cultures growing at 37˚C at 1 µL/mL. After being kept overnight, a second round of IPTG, glucose, and MgSO4 was added.
Protein characterization
Cell lysate was isolated into insoluble and soluble fractions; both samples were then run through a Lumio™ gel (using a Lumio™ Green Detection Kit, ThermoFisher Scientific, Waltham, MA, USA) with luminescent ladders to confirm presence of DXS synthase and SRPP. Observed bands were then compared to the expected band size of all proteins to confirm results.
Biogas production
C. reinhardtii strain 90 was ordered from UTEX (205 W. 24th St, Biological Labs 218, Austin, TX 78712) in glass tubes filled with agar. All cultures for testing were grown from this original stock. To begin, C. reinhardtii was scraped from the original agar stock and added it in 10 mL sulfur-free BG-11 media, with exposure to light at 21 W/m2 in 12 hour cycles. BG-11 was chosen for being a commonly-used medium for growing freshwater algae [16].
Membrane protection
Melanin
The UV protective properties of melanin make the pigment an ideal candidate for increasing radiation resistance in a balloon membrane. Eumelanin synthesis is initiated by a copper containing tyrosinase acting on the aromatic amino acid L-tyrosine. Tyrosinases catalyze the hydroxylation of L-tyrosine to L-DOPA, as well as a further oxidation to produce the cyclical dopachrome. Eumelanin is formed in vivo by the non-enzymatic oxidation and polymerization of dopachrome [17]. The MutmelA gene endogenous to Rhizobium etli encodes a feedback resistant tyrosinase, shown to be effective when transformed into E. coli [18]. L-tyrosine, along with many other common aromatic compounds, is produced endogenously in E. coli through the shikimate pathway. To increase intracellular L-tyrosine concentrations in E. coli, carbon can be shuttled from the cells’ central metabolism into the shikimate pathway [18]. DAHP synthase catalyzes the conversion of Phosphoenolpyruvic acid (PEP) and Erythrose 4-phosphate (E4P) to DAHP in the shikimate pathway’s initial reaction. PEP and E4P are naturally derived from glucose, and are present in the cytoplasm of E. coli grown in glucose containing medium. We aimed to decrease metabolic consumption of PEP and E4P to increase the flux of DAHP synthase. PEP and E4P are consumed by the glucose phosphotransferase transport system (PTS). Inactivation of this system has been shown to increase the yield of these compounds in E. coli grown in a glucose medium [18]. We acquired a previously published PTS- tyrR- glucose+ strain VH33tyrR [18], derived from W3110 E. coli contained genomic knockouts of PTS and the tyrR negative transcriptional regulatory protein, decreasing the cellular consumption of PEP and E4P and increasing the biosynthesis of and transport of tyrosine.
Three isoenzymes of DAHP synthase are endogenous to E. coli, each inhibited by different amino acids produced through the pathway. This negative feedback limits the production capacity of L-tyrosine in wild type E. coli. Cultures of the VH33tyrR strain were transformed with plasmids containing feedback resistant forms of the AroG and AroF isozymes, as well as the MutmelA gene to stimulate melanin production. Cells were transformed using 2×TSS made with LB broth containing 10% PEG, 5% DMSO, and 50mM Mg2+.
Sensing with chromoproteins
To create a biological temperature sensor, chromogenic proteins were obtained from the ATUM (Newark, CA) ProteinPaintbox and from the iGEM registry.
Plasmid assembly
To investigate the changes undergone by these chromoproteins when heated, coding sequences for AE Blue and 12 ATUM chromoproteins (PrancerPurple, CupidPink, TinselPurple, VixenPurple, MaccabeePurple, SeraphinaPink, LeorOrange, ScroogeOrange, BlitzenBlue, DreidelTeal, DonnerMagenta, VirginiaViolet) were cloned into pSB1C3 with Gibson Assembly. A FLAG, lumino and 6x histidine tag were added by flanking primer ends in order to extract the expressed protein using nickel column purification. The lumino tag is a specific six amino acid sequence that binds to LumioTM Green [19] which allows the fusion of the lumino tag and the chromoprotein to be detected on an SDS-PAGE gel without having to run a staining protocol. The FLAG tag allows for anything after its sequence to be cleaved off of the protein when the extracted protein is incubated with enterokinase [20], in case the lumino or histidine tag interferes with the chromoprotein structure.
Additionally, a cellulose-binding domain isolated from PCR was added to each of the chromoproteins. The cellulose binding domain sequence isolated from Cellulomonas fimi has been shown to bind irreversibly to cellulose [21]. In order to ensure that the addition of this larger protein did not interfere with the structure of the chromoproteins, both chromoprotein versions without the cellulose binding domain and with the domain were tested.
Protein Purification
After performing a Gibson Assembly to place the DNA sequence for the ATUM chromoprotein and the sequence for the Flag-Lumino-His Tag, all twelve of the ATUM chromoproteins were color-verified via colony PCR using iGEM’s VF and VR primers (S1 Fig). Next, a large (250-500 mL) liquid culture for each of the cells expressing the chromoprotein then extracted the expressed protein using 3 mL HisPur Ni-NTA nickel columns (ThermoFisher Scientific, Waltham, MA, USA). The extracted protein was then dialyzed and concentrated using microfiltration centrifuge tubes in order to perform heat tests. An apparatus was used that would heat a glass petri dish evenly and allow for a video to be taken of the cell lysate as it was being heated.
Lyophilization
In order to investigate the role that water molecules play in the chromoproteins’ color change observed in response to heat, a lyophilizer vacuum was utilized to sublime the water from the chromoproteins without heating them. The water vapor was then pumped out of the system. After an hour in the lyophilizer, the filter paper was removed from the chamber and photographed. Water was then added to the right column of the filter paper which caused the color to return.
Sensing with aptamer probes
A fluorescent molecule (fluorophore) was synthesized directly onto the 5’ end of an ATP aptamer with a biotinylated 3’ end. From there, a complementary oligonucleotide to the 5’ end of the aptamer was designed, with a quencher added to the end. When both strands were allowed to incubate in a Streptavidin-coated plate, a complex was created where both oligonucleotides bound together to form a quenched sensor complex that is attached to the Streptavidin-coated surface. When target ligand is introduced, the aptamer undergoes conformational change as it folds around the ligand. This conformational change sterically displaces the quencher oligonucleotide and exposes the fluorophore, thus emitting detectable signal [22].
To couple the biosensor to a proof-of-concept membrane, we purified the 2014 Stanford-Brown-Spelman iGEM team’s part BBa_K1499004 (validating the presence of its HisTag) and used their cellulose cross-linker to bind a fluorophore sensor, with quencher. This incubated concoction was then distributed to wax-printed cellulose filter paper to measure the binding activity to the paper over a week. Initially, the mixture was applied to the paper and incubated 2-3 days for cellulose binding. After this initial binding period, 5 x 1 mL of milliQ water (with 1mM ATP) were washed over each 9-well sample each day for a week. The positive control had the FQ system, but no linker. The negative control had neither the FQ system nor the linker.
Results
Latex
Engineering E. coli to produce cis-1,4 polyisoprenes (latex)
In order to form our bio-membrane, we designed a transgenic organism capable of mimicking the natural rubber production process found in H. brasiliensis [23]. Not only could our transgenic single cell organism allow for optimization of the polymer synthesis pathway, but also could be grown to yield a large volumes cis-polyisoprene (synthetic rubber) polymers with minimal growth media input.
Rubber (cis-1,4-polyisoprene) synthesis pathway
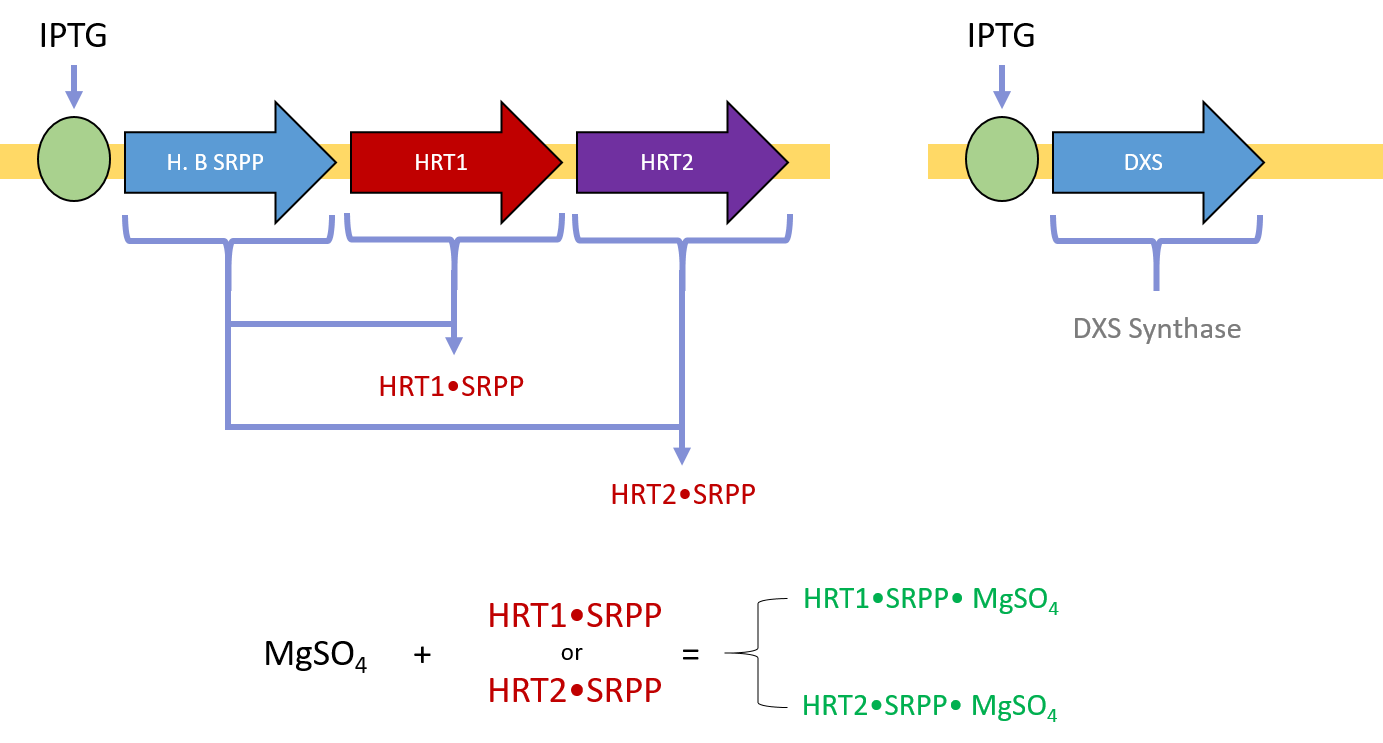
Although there are a variety of polymer forms, natural rubber is largely composed of cis-1,4-polyisoprene. To produce cis-1,4-polyisoprene, rubber trees such as H. brasiliensis express cis-prenyltransferase enzymes (rubber transferases) to link individual isoprene monomers into a polymer. The primary monomer in cis-1,4-polyisoprene is isopentenyl pyrophosphate (IPP), which has a more reactive isomer, dimethyl-allyl pyrophosphate (DMAPP) [24, 25]. To initialize chain elongation, prenyltransferase enzymes start with a single DMAPP molecule and iteratively extend the chain with IPP. However, prenyltransferase enzymes require a magnesium (II) supplement and small rubber particle protein (SRPP) coenzyme for activation [24]. To polymerize rubber chains, the key xenogeneic components our host organism will need include (1) prenyltransferases, (2) IPP substrate, (3) DMAPP substrate, (4) a magnesium (II) supplement (such as MgSO4), and (5) small rubber particle protein coenzymes.
DXS synthase optimization
Although IPP and DMAPP could be supplied directly to the media, producing both within the host organism from a more available precursor would reduce cost of synthesis. IPP and DMAPP are endogenous to many species, and two common IPP biosynthesis pathways were identified: the mevalonate (MVA) pathway and methylerythritol phosphate (MEP/DOXP) pathway. While both pathways generate IPP and DMAPP from sugar, only the MEP/DOXP pathway is endogenous to E. coli [26]. Through a six step process, the MEP/DOXP pathway converts pyruvate and glyceraldehyde 3-phosphate (G3P) into IPP and DMAPP (Fig 1). While incorporating the MVA pathway into E. coli would further enhance IPP and DMAPP output, we opted for optimizing the MEP/DOXP pathway to minimize the step count from converting a substrate into IPP [27, 28]. Additionally, since G3P and pyruvate are products of glycolysis, providing a glucose supplement to E. coli would fuel G3P and pyruvate use by the MEP/DOXP pathway [29].
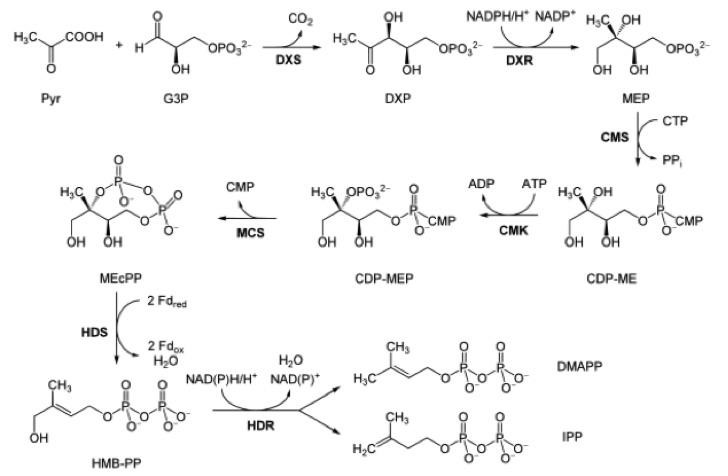
The sequence for the DXS gene was obtained from EcoGene (EG13612). For future protein purification and characterization, a FLAG, tetracysteine, and hexahistidine tag were attached to the DXS gene. A double terminator, (BBa_B0010 and BBa_B0012) was used to ensure appropriate transcriptional arrest [31]. The construct was synthesized in two parts, and Gibson assembly was used to insert them into linearized pSB1A3. The resulting construct was transformed into T7 express cells.
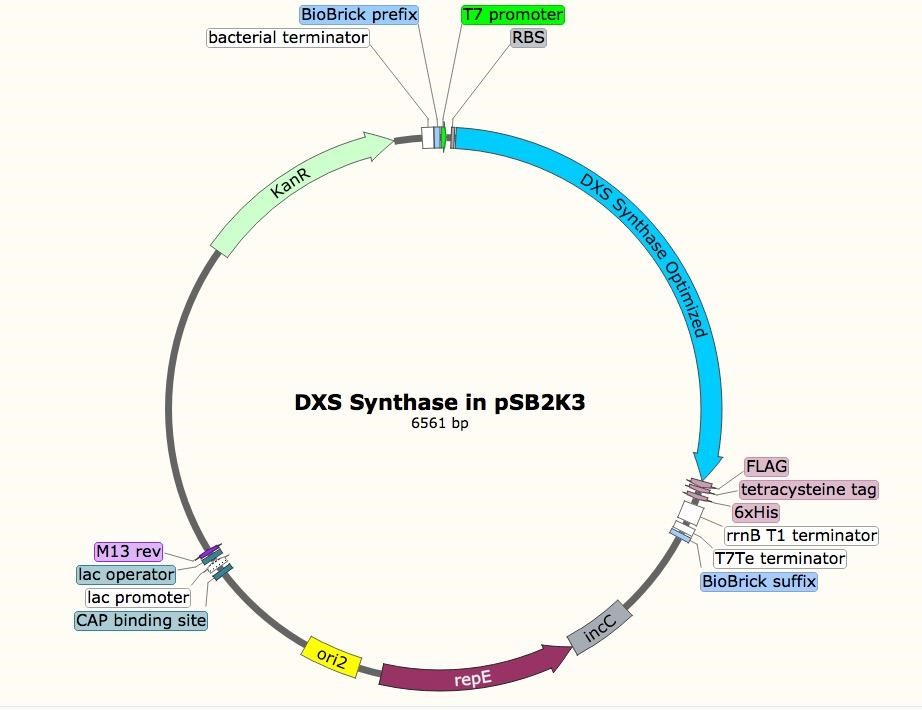
Incorporating rubber synthesis into E. coli
While DXS synthase optimization can increase the amount of IPP and DMAPP substrate, we also needed to incorporate the isoprene synthesis pathway from H. brasiliensis into E. coli. We identified well-characterized cDNA sequences from H. Brasiliensis for prenyltransferases directly involved in natural rubber biosynthesis. Two enzymes were selected, HRT1 and HRT2 (Accession No. AB061234.2 and AB064661.2), which are two cis-prenyl chain elongating enzymes isolated from Hevea latex [32]. Found predominantly in fresh Hevea latex, these proteins synthesize new rubber molecules. Both genes were incorporated into a plasmid under an IPTG-inducible promoter.
Since prenyltransferase requires an SRPP and various chemical cofactors for enzymatic functionality, we identified the most commonly expressed SRPP in Hevea latex as a potential cofactor for HRT1 or HRT2 [33, 34]. However, because there are different classes of SRPPs that can play a variety of roles in Hevea latex production, selected SRPPs also were identified as rubber elongation factors [35]. Small SRPPs were also selected over large rubber particle proteins, as the latter would be difficult to produce in E. coli due to their size [36]. The gene of the single SRPP (AF051317) was subsequently incorporated into the prenyltransferase cassette [27]. Since prenyltransferase activity depends on SRPP presence and gene order in an operon can influence expression level, the SRPP gene was inserted before the two prenyltransferases. This cassette was then linked to IPTG-inducible T7 Elowitz high-copy promoters to allow for regulated expression. All proteins were also tagged with a FLAG, tetracysteine, and hexahistidine tag to allow for protein purification assay (Fig 3).
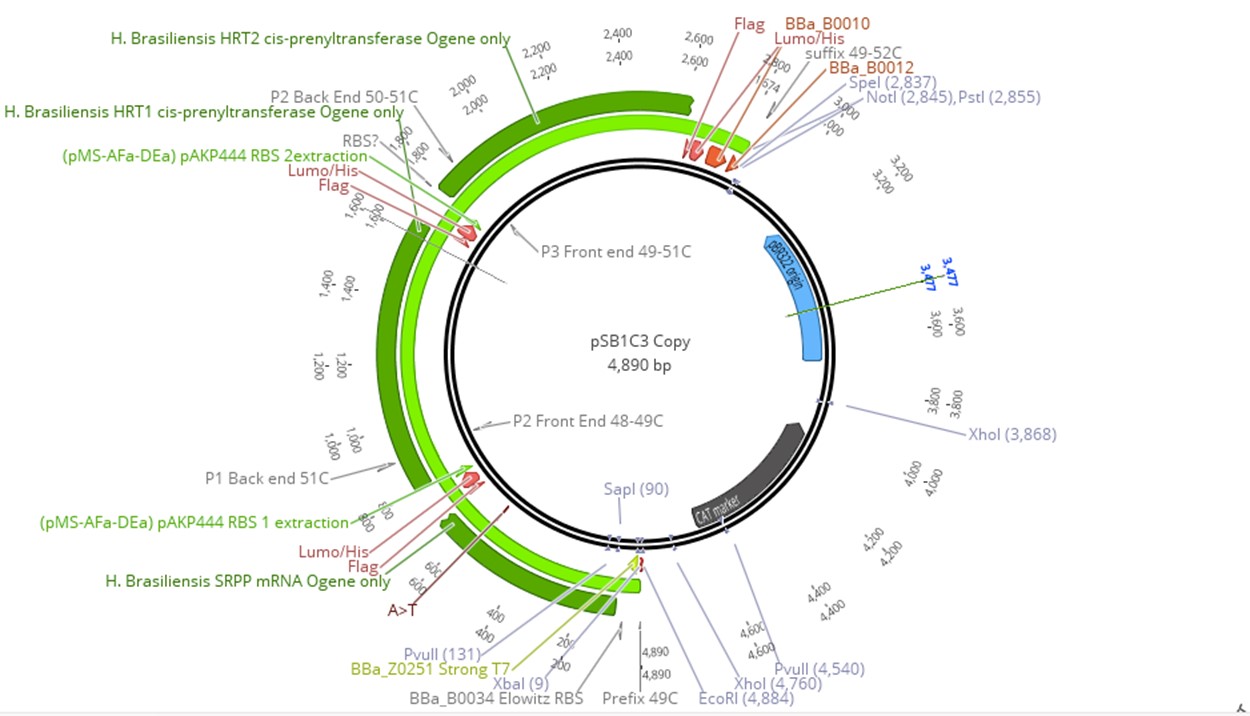
To produce our plasmid, SRPP, HRT1, HRT2 were synthesized independently and incorporated into pSB1C3 using Gibson Assembly. The construct (latex operon) was then transformed into T7 express cells. Although our system was incorporated on two plasmids, all four components (HRT1, HRT2, SRPP, and DXS synthase) could be included on the same construct for ease of transfection. However, transformation of E. coli with the DXS plasmid would allow for an organism that produces IPP and DMAPP, which could be useful for terpenoid synthesis. For full realization of high throughput IPP and cis 1,4-polyisoprene production in E. coli, all plasmids were transformed into the host to allow for high expression of all pathway elements. To verify all elements were property transformed, each plasmid had distinct selection markers. Due to similarity in ORI between pSB1C3 and pSB2K3, the latter was replaced with pUC19 for the backbone.
After E. coli cells have been transformed with both plasmids, a supplement of magnesium sulfate, IPTG, and glucose is sufficient to induce protein expression, activity, and cis-polyisoprene polymer synthesis. HRT1 and HRT2 bind with SRPP and Mg(II) to become enzymatically activated (Fig 4). DXS synthase accelerates the conversion of pyruvate and G3P to DXP. The activated HRT1 and HRT2 complexes can then extend IPP into polyisoprene via dephosphorization (Fig 5).
Latex extraction was performed on 350 ml of our dually transformed cells containing the plasmids for the Latex Operon and DXS Synthase. Cells were pelleted and then lysed with chloroform to dissolve polyisoprenes in solution [37, 27]. The solution then filtered to yield chloroform and its solutes. Methanol was then added to precipitate the latex from the chloroform solution [37]. The LB fraction was also subject to a similar procedure, except the chloroform layer was obtained by separation of the solvent layers.
After this procedure, 1.5 ml of a rubbery, off-white polymer from the methanol layer precipitated (Fig 6). For a quick characterization test, the polymer was dried and burned, which resulted in a smell of burning rubber and a strong black smoke—typical of rubber fires and identical to our control test where a sample of latex was ignited [38]. Additionally, our dried extract is highly compressible and elastic. Further tests with mass spectrometry, acetone characterization, degradation, and physical stress tests will be needed to confirm the composition of the polymer. Questions we hope to answer with continued development are whether our product can be used as a rubber substitute in industrial applications, steps that could be taken to optimize for product yield, and whether our product could be tagged for secretion into cell culture media.
Membrane protection
Melanin Binding Peptides
Coating high altitude balloons with any substance can introduce inconsistencies in the membrane’s tensile properties. With elastic membranes, balloon expansion at different altitudes can expose weak spots susceptible to tearing, thereby shortening the balloon’s lifespan. To mitigate these issues, we explored methods to incorporate melanin directly into our biological membranes. Studies investigating the use of melanin as a target molecule for localized radionuclide therapy of melanoma cells have utilized phage display libraries to identify a set of short melanin binding peptides [39, 40]. Identifying the findings with the highest melanin retention rates, we decided to pursue the decapeptide known as 4B4 and a hepapeptide called 4D, along with control peptides PA1 and P601G for reference. Qualitative UV properties were assessed using the NASA Ames Solar Simulator (1300W of power provides radiation comparable to UV bursts during solar flares to simulate the harsh conditions of space). The results can be described on the 2016 Stanford-Brown iGEM Wiki.
Repetitive nucleotide sequence assembly
To fix as many melanin molecules to a single protein binding agent, we designed genetic constructs coding for a repetitive sequence of melanin binding peptides connected via a GS linker to a cellulose binding domain. We looked into three different test constructs to achieve optimal melanin localization: two homogeneous repeats of 4B4 and 4D respectively, and one randomized heterogeneous assembly of 4B4 and 4D repeats (S2 Fig). To avoid errors inherent to DNA synthesis of long strands containing many repeats, we used a newly developed overlap based DNA block assembly and shuffling method [41]. Codon optimized single stranded oligonucleotides for E. coli were synthesized that coded for 4B4, 4D, PA1, and P601G, each with identical 3′ and 5′ six nucleotide sticky end overhangs. By phosphorylating the DNA blocks’ 5′ ends with T4 Polynucleotide Kinase (NEB), and allowing the strands to ligate overnight with T4 DNA Ligase (NEB), we were able to generate up to 30 repeated segments per strand (S3 Fig). Elongation is halted when a DNA block contains a deletion, insertion, or substitution on its 5′ overhang, which prevents further DNA blocks from annealing to its sticky end. Repetitive 4D and P601G strands were assembled behind constitutive promoters and strong RBSs into pSB1C3 backbones. Downstream of the genes, a cellulose binding domain (CBD) extracted from the 2016 iGEM Distribution Kit (part BBa_K1321357) was included as a proof of concept membrane binding domain, separated from the melanin binding module by a GS linker to preserve each domain’s function. Following the CBD, a FLAG tag, Lumino tag, and His tag were added for protein presence verification, extraction, and purification. Finally, a double terminator was included before the biobrick suffix to insure termination of transcription.
Biogas production
Generally, C. reinhardtii absorbs CO2 from the atmosphere to create various carbohydrates, releasing oxygen gas as a byproduct [11]. Green algae in particular can produce hydrogen gas instead of oxygen through direct biophotolysis [42], where a reversible hydrogenase enzyme catalyzes a reaction between photosystem II and ferredoxin. The water-splitting reaction of photosystem II creates electrons that are sent to ferredoxin, and the reversible hydrogenase in the stroma of the chloroplast combines these electrons with free-floating protons in the medium to create H2 [43]. C. reinhardtii’s reversible hydrogenase is extremely sensitive to O2 pressure, and will irreversibly inactivate upon sensing oxygen. Thus the alga’s photosynthetic production of H2 and O2 must be temporally separated, and can be done upon inducing two-stage direct biophotolysis. CO2 is fixed normally through oxygenic photosynthesis during Stage 1, and H2 is then generated under anaerobic conditions in Stage 2. This can be done by depleting sulfur from the culture medium [44].
Prior research by Jo et al. (2006) [45] has shown that optimizing growth conditions for C. reinhardtii can result in a little over 2 mL of H2 produced for a 10 mL culture of alga after 96 hours. This optimized process gave a hydrogen production rate approximately 1.55 times higher than typical cultivation in sulfur deprived TAP medium. Our process was not perfectly optimized, so we expected a gas production rate of approximately 1.33 mL of H2 for a 10 mL culture of algae after 96 hours. After the algae had grown to a sufficient density, the stock of C. reinhardtii was allowed to fill a flask with gas, and then conducted a simple chemistry test to see if we had been producing oxygen or hydrogen [46]. Our expectations were as follows: when introducing a lighted splint to our flask, contact with hydrogen gas would result in a loud noise, alongside the extinguishment of our flame. Contact with oxygen gas should produce the opposite effect of re-lighting our flint or strengthening the flame, as oxygen gas supports combustion. Upon testing our splint with our flask of algae, a noise was heard and our flame was extinguished, suggesting the presence of hydrogen gas.
Sensing with chromoproteins
An assortment of 12 chromoproteins (PrancerPurple-PP, CupidPink-CP, TinselPurple-TP, VixenPurple-VP, MaccabeePurple-MP, SeraphinaPink-SP, LeorOrange-LO, ScroogeOrange-SO, BlitzenBlue-BB, DreidelTeal-DT, DonnerMagenta-DM, VirginiaViolet-VV; provided by ATUM, Newark, CA) were tested in a closed system with no air interaction.
Cell lysate heat tests
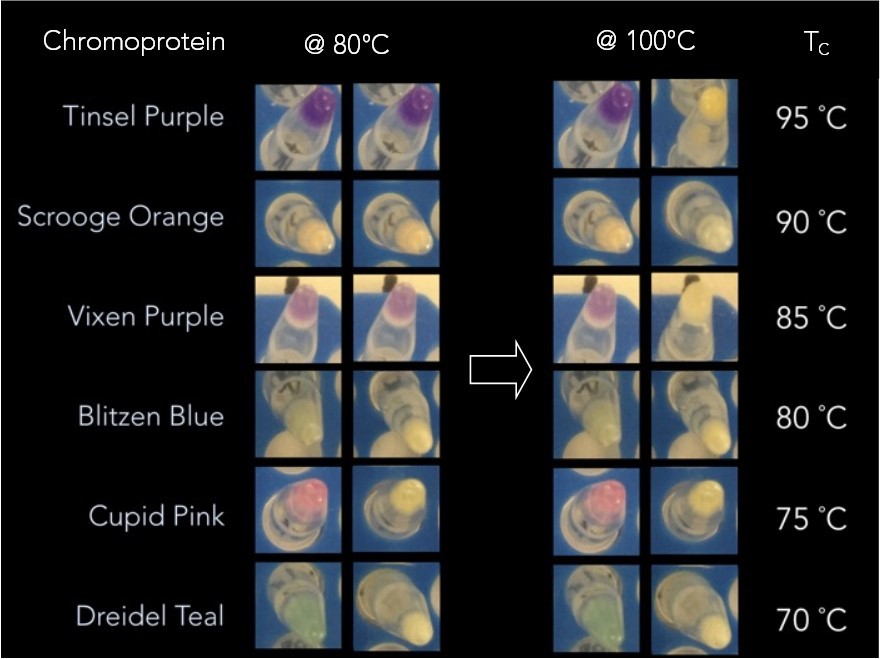
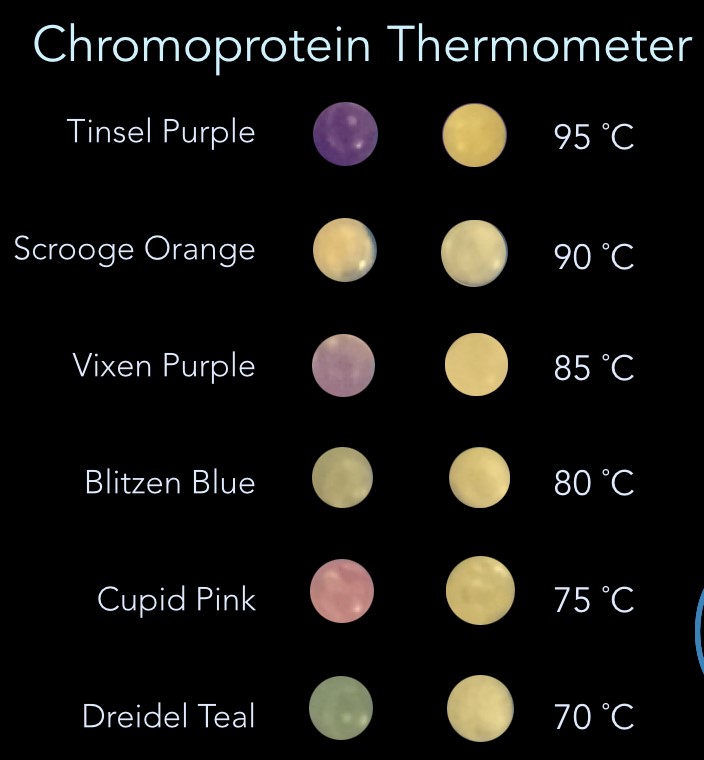
5.0 mL cultures of each chromoprotein were spun down and 30 µL of the pellet cells with 1 µL of 0.165 M EDTA were added. The PCR tubes were then heated for 5 min at a starting temperature of 40 °C and removed from the thermal cycler. A picture was taken before returning them to the thermal cycler for 5 more minutes at a temperature 5 °C higher than the previous. This process was repeated until 100 °C was reached. As can be seen in Fig 7, groups of chromoproteins lose their color at specific temperatures. Additionally, CupidPink and DonnerMagenta change from pink to purple at 70 °C and 80 °C, respectively.

Using the results of the experiment above, it was possible to build a thermometer from the select groups of chromoproteins that respond to different temperatures. Fig 8 shows a prototype of how that would work at 80°C (left) and 100°C (right). Until the ambient temperature reaches the temperature indicated on the temperature scale on the right, the color remains constant. When it exceeds that temperature, color is lost or significantly changed.
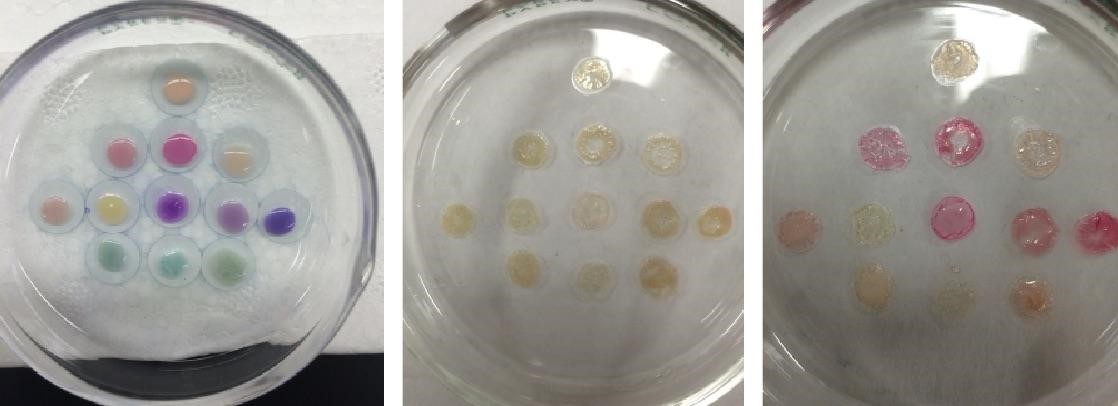
Twelve chromoproteins were selected for a heat test assay and can be seen in Fig 9 on the left. These cell lysate heat tests were done in a closed environment, so further tests were done in an exposed air environment. This was based on the idea that the observed color change was dependent on the interaction of water molecules with the barrel of the chromoproteins [13]. In the exposed air system, the water molecules would not be trapped with the chromoproteins and color change might be observed sooner. Chromoproteins were spotted on a glass petri dish heated to 85 °C, which caused all of the chromoproteins to lose color as seen in the center panel of Fig 9. The glass petri dish was then removed from heat and 20 µL of DI water added to each of the chromoproteins (Fig 9, right). If the color loss was due to loss of water molecules and not to denaturation, this rehydration step should restore color.
Hydration assay results
When testing the cell lysate of the chromoproteins in a closed system it was observed that color expressions were sometimes variable, but low heat treatments performed in the closed system described above caused color to appear. Five chromoproteins from ATUM were tested: VirginiaViolet, MaccabeePurple, SeraphinaPink, TinselPurple, and DonnerMagenta. Five PCR tubes containing 30 µL of the cell pellet do not show any color. After these previously colorless cell pellets have been treated at 55 °C for 30 min, 60 °C for 30 min, and 60 °C for 30 min, color could be observed. This color then proceeded to disappear again as the temperature continued to increase, and the color did not return after subsequent heat treatments.
As can be seen above in Fig 9, some of the chromoproteins regained color when water was added, but this color is different from the starting color. This could be due to the fact that the heating caused the chromoprotein structure to destabilize to a lower energy structure which causes it to have a different color when the water is added to support the barrel. These results show that while color loss does occur when water is removed from the system, it does not have the same effect as heating the sample. In the heat testing experiments, PrancerPurple changed irreversibly to pink when heated whereas in the lyophilization experiment, it regains its color fully. This supports our hypothesis that the color changing in response to heat is dependent on both water loss and a structural change of the protein itself due to heat.
Similar experiments were done with the AE Blue-CBD fusion protein. With this protein, we saw that the blue color changed from blue to purple. This purple color immediately went away when the sample was removed from the chamber, suggesting that our hypothesis that the AE Blue chromoprotein’s color changing capabilities are dependent on the presence of water inside the barrel. To further confirm this, we tested how the AE Blue-CBD fusion protein would respond to heat in a closed PCR tube. Since the PCR tube prevents water from being released from the system, the water molecules that support the barrel of the chromoprotein cannot be driven out when the tube is heated. Because of this, we no longer see the color change from purple to blue. Instead the chromoprotein turns a blue-green color, likely indicative of the protein simply denaturing in response to heat (Fig 10).
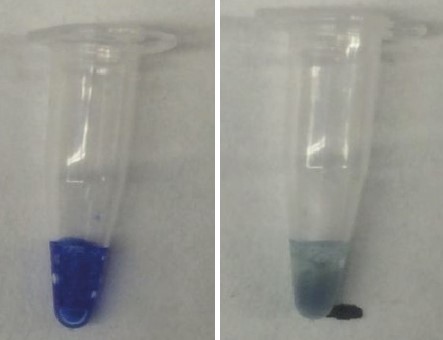
Water molecules are essential for the chromogenic protein to maintain its structure. In response to heat, our hypothesis is that these water molecules are released from the beta barrel, thus allowing the barrel to collapse and the chromophore to no longer be supported enough to produce color. The extracted 12 ATUM chromoproteins with FLAG lumino his-tags were concentrated using microfiltration tubes then allowed to dry on cellulose sheets which had wax wells printed on them. These cellulose sheets with the dried chromoprotein were then placed in a preheated oven and the temperature was increased at five minute intervals until a temperature change was observed (Fig 11). This test shows a reversible color change for all 12 chromoproteins at lower temperatures than seen in cell lysate tests and also shows this heat testing can be done multiple times with consistent results. Because of the range of temperatures the chromoproteins lose color at, there is the application for a paper based thermometer.

Prototyping a biological thermometer
In the interest of moving towards a biologically based paper thermometer, a Gibson Assembly was performed to add a cellulose binding domain to the end of the chromoproteins. This was first done successfully with AE Blue from the iGEM registry, then with the 12 ATUM chromoproteins. Once this construct was successfully transformed and expressed in bacteria, the protein was extracted and dialyzed. The concentrated protein was then pipetted onto cellulose sheets and underwent a series of heat tests. It was found that, while this protein initially appeared blue, (Fig 12, left) it begins to turn purple at 55 °C (Fig 12, center left) and this purple color becomes more pronounced as temperature is increased (Fig 12, center right). When allowed to cool back to room temperature, the blue color returned (Fig 12, right).
A sticker was then created using the AE blue fusion protein to coat a cellulose sheet. This sticker was then fixed to a glass container using double sided tape (Fig 13, left). Hot water was added to the glass container and after 1 minute of exposure to heat, the AE blue-CBD fusion protein turned purple (Fig 13, center). Cold water was then added to the glass container and the AE blue-CBD fusion protein returned to its original blue color (Fig 13, right). This testing shows a reversible, color changing, temperature sensitive fusion protein and is the prototype of a biologically-based paper thermometer.

To further test the binding efficacy of our cellulose binding domains, we introduced two chromoproteins that had been given cellulose binding domains (PrancerPurple and LeorOrange) to cellulose sheets. After being allowed to set for three days, the binding capabilities of the cellulose binding domain was tested by washing the cellulose sheet the chromoproteins were bound to with a steady stream of water for 1 minute. After this washing, no color change was detected, indicating that the cellulose binding domain was effective at fixing the chromoproteins to the sheet.
Sensing with aptamer probes
To validate the results of Nutiu & Li 2005 [22] and measure the sensitivity and specificity of our functionalized ATP aptamer, we developed an assay comparing the sensing of various NTP and dNTP’s. Fig 14 shows the data collected in testing the biotinylated ATP FQ sensor bound to a streptavidin plate without a cellulose cross-linker (data for controls unattached in solution and with cross-linker can be found on the Stanford-Brown iGEM Team Wiki: http://2016.igem.org/Team:Stanford-Brown/SB16_BioSensor_FQsensor). Fluorescence was measured using the SpectraMax GeminiXS Fluorescence Plate Reader and normalized against the negative control well for a fold-ratio.
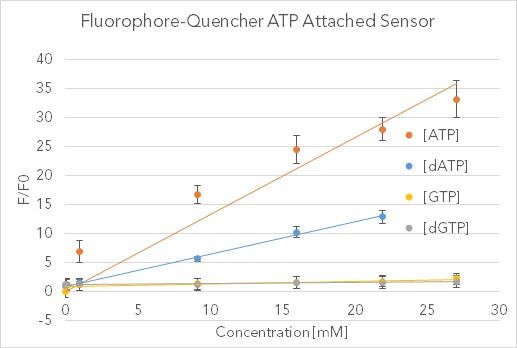
After validating the sensor’s fluorescent properties in solution with 30-fold increases relative to similar nucleotide triphosphates, we sought to integrate this sensor into a biodevice with the Stanford-Brown-Spelman iGEM team’s part BBa_K1499004. Over the course of a week, fluorescent images were analyzed with Typhoon Scanner (Absorption = 495 nm, Emission = 525 nm) and processed quantitatively with Python. This washing assay provided qualitative proof that both the FQ sensor and Cellulose Cross-linker are working in tandem, as evidenced by the drastic difference in fluorescence between the experiment and controls (see http://2016.igem.org/Team:Stanford-Brown/SB16_BioSensor_FQsensor). Photobleaching and heterogeneous mixing across the wells are uncontrolled for, but this pilot study confirms the possibility of embedded abiotic nucleic-acid based sensors.
Discussion
Membranes
Based on our results, the material that could yield further interesting experimental results is the synthetic rubber produced through our manipulation of E. coli. Through metabolic engineering we produced an organism capable of producing polyisoprenes. By optimizing the endogenous MEP/DOXP pathway in E. coli through overexpression of rate-limiting proteins and coupling it with expression of rubber transferases (prenyltransferases) used by H. brasiliensis for polyisoprene production, we obtained a rubber-like substance that exhibits the basic properties of latex. Further tests with mass spectrometry, acetone characterization, degradation, and physical stress tests will be needed to confirm the composition of the polymer. Questions we hope to answer with continued development are whether our product can be used as a rubber substitute in industrial applications, steps that could be taken to optimize for product yield, and whether our product could be tagged for secretion into cell culture media, mitigating the need for cell lysis for recovery of the product.
Because our initial ignition and mechanical property tests are not indicative of whether or not our product is chemically identical to cis 1,4 polyisoprene, additional chemical tests need to be performed. An H1NMR and C13NMR assay of our extract would allow for better characterization of the chemical properties of our extract. Comparison of H1NMR and C13NMR peak placement with pure polyisoprene will be indicative of the similarities and differences in chemical composition of our product with actual synthetic rubber. Additionally, because our manipulation of the MEP/DOXP pathway and HRT1, HRT2 enzyme activity is not entirely optimized, future studies will include further optimization of the MEP/DOXP pathway and directed evolution of HRT1, HRT2 protein constructs to allow for faster production times. Furthermore, our yield is currently 1-2% by volume; this return could be further enhanced by adding an export tag to polyisoprene compounds that could be later cleaved. This would not only allow for more polyisoprene compounds to be dissolved in solution and easier to purify, but also prevent the host cell from using polyisoprenes as an energy source.
Sensing
Our chromoprotein research not only characterized the way in which chromogenic proteins lose or change colors in response to heat, but also was expansive enough to identify a large span of chromogenic proteins that differentially respond to temperature. Using this differential response, a biological thermometer was constructed that was made up of six different chromogenic proteins. Each of the proteins in this set loses or changes color at a specific and distinct temperature. Lastly, the addition of a cellulose binding domain to these chromogenic proteins to allow them to bind to cellulose, allowing for the creation of a biologically-based paper thermometer.
From our FQ sensor data, we concluded that this sensor worked well with high ATP concentrations, and with further research could almost certainly be optimized to sense as low as nanomolar concentrations. Advantages of this system include its customizability (theoretically it should work to detect any molecule for which there exists an aptamer) and its ability to retain sensory functionality while bound to a solid surface. Disadvantages include only detecting high concentrations of target ligand (at least currently, though this could theoretically be at least somewhat mitigated with further efforts at system optimization), and the fact that this system is fundamentally synthetic, limiting its ability to be easily and cheaply replicated in vivo like other bio-bricked parts. Instead, expensive new constructs have to be ordered to build each new sensor. In our qualitative and quantitative tests, the FQ sensor with the cellulose linker performed as well as or better than the FQ sensor without the linker. This suggests that our FQ sensing platform may work in conjunction with other membrane cross-linkers, and it may provide a path forward in developing membrane-attachable small-molecule sensors.
For both sensors in remote settings, a major obstacle remains imaging the throughput. While alternative biological outputs such as electron chains or proton donation could be integrated into electrical sensors, using these light and fluorescence-based sensors on a large, replicated scale may suffice in conjunction with an orbiting satellite to scan the sensor data.
Membrane Protection
The results described herein provide qualitative confirmation of increased UV resistance. UV-Vis absorbance spectroscopy analysis of the runoff generated from a wash of cellulose sheets coated in a mixture of melanin and our novel melanin-cellulose linker demonstrated an increased retention rate of bound melanin. These findings support the alternative approach of including protective agents directly into the balloon membrane rather than as a coating. A molecular binding agent such as the construct discussed above could help mitigate effects of balloon coatings such as heterogeneous tensile properties due to uneven coating distribution. Furthermore, a binding agent is necessary to incorporate protective materials into elastic membranes to accommodate an increased surface area in response to balloon expansion at different altitudes. This melanin binding construct could be integrated with natural rubber latex or an elastin/collagen hybrid membrane to increase the lifespan of biodegradable balloons. The tradeoff in mass necessary to provide enough UV protection may limit the maximum altitude of balloons using this technology; however, dealing with the radiation is an unmet need in both ballooning and space applications, so these results can have promising implications for further research.
Biogas
The biogas experiment was a necessary validation for our proof-of-concept, as all components of the desired BioBalloon must be produced biologically. In this sense, we accomplished validation goals for this subproject and utilized a commonly-found algae to produce both hydrogen and oxygen gas. In a BioBalloon prototype, it is possible for C. reinhardtii to be kept in contained quarters in space, while producing gas that is then injected into the BioBalloon so that it can take flight in extraterrestrial atmospheres.
Conclusions
The work done here can be built on in the future to produce a functional prototype for a balloon made out of biological products, while at the same time advancing a number of smaller projects worthwhile in their own right. In particular, production of an elastic material from genetically engineered E. coli culture is potentially indicative of a novel implementation of recombinant latex production, and regardless of the chemical identity of the product represents a successful protocol for recombinant production of an elastic material. Tuning this protocol in particular in the future with the advantage of more detailed study may result in increased yield, and as metabolic engineering is an active field of study the economics of recombinant material production continue to evolve. Select chromoproteins have been shown to be visibly sensitive to temperature while remaining robust to careful addition of “adapters,” using cellulose binding domains as a proof of concept for producing functional membranes made of biological materials. While at the current stage “growing” a functional balloon in a microbiology lab is not possible, the relevant raw materials can be produced in the lab. Even before these individual pieces might be refined and combined into a ‘BioBalloon,’ each project can be pursued independently based solely on its individual merits. Focusing on just two examples, the utility of chromoproteins as lab indicators necessitates reliable and thorough characterization in any case, and rubber from any source has myriad uses beyond balloon construction. The independent value of each component ensures tangible results at intermediate stages of a complex project, an approach which mitigates the risks of undertaking what must necessarily be a multi-year project.
Acknowledgments
The authors would like to thank the lab of Dr. Lynn Rothschild, including Dr. Kosuke Fujishima, Dr. Ivan Paulino-Lima, Dr. Mark Ditzler, Griffin McCutcheon, Ryan Kent (Brown-Stanford iGEM 2011), Jesica Navarrete, Simon Vecchioni (Stanford-Brown iGEM 2013). Dr. Kara Rogers provided guidance and mentoring at Stanford, Professors. Gary Wessel and James Head at Brown, and Jill Tartar, Margaret Race, Maria Chavez. We are indebted to ATUM for their contribution of chromoproteins from their ProteinPaintbox, and the Stanford Space Initiative for providing a flight platform to test them. We would also like to thank the remaining team members, Julia Gross and Taylor Pullinger, for their technical assistance.
Response to Reviewers
A transcript of the reviewer comments and author responses from the Live Peer Review Jamboree can be found here: Stanford-Brown Response to Reviewers
References
- Gold MN. The Wrong Stuff: America’s Aerospace Export Control Crisis. Neb. L. Rev.. 2008;87:521.
- Herrmann KM. The shikimate pathway: early steps in the biosynthesis of aromatic compounds. The Plant Cell. 1995 Jul;7(7):907.
- Bank AJ, Wang H, Holte JE, Mullen K, Shammas R, Kubo SH. Contribution of collagen, elastin, and smooth muscle to in vivo human brachial artery wall stress and elastic modulus. Circulation. 1996 Dec 15;94(12):3263-70.
- Burnham, Michael. “Can Space Travel Be Environmentally Friendly?” Scientific American. Scientific American, 27 May 2009. Web.
- Kramer, Sarah and Mosher, David. Here’s how much money it actually costs to launch stuff into space. Business Insider: Science. Jul. 20, 2016. Web.
- World Butadiene Supply to Tighten [Internet]. Oil and Gas Journal. Oil and Gas Journal; 2001 [cited 2017Mar5]. Available from: http://www.ogj.com/articles/print/volume-99/issue-4/processing/world-butadiene-supply-to-tighten.html
- Li H, Aide TM, Ma Y, Liu W, Cao M. Demand for rubber is causing the loss of high diversity rain forest in SW China. Biodiversity and Conservation. 2007 Jun 1;16(6):1731-45.
- Miller AJ, Tsao H. New insights into pigmentary pathways and skin cancer. British journal of dermatology. 2010 Jan 1;162(1):22-8.
- Brenner M, Hearing VJ. The protective role of melanin against UV damage in human skin. Photochemistry and photobiology. 2008 May 1;84(3):539-49.
- Williams DR. Mars Fact Sheet [Internet]. NASA. NASA; 2016 [cited 2017Mar5]. Available from: https://nssdc.gsfc.nasa.gov/planetary/factsheet/marsfact.html
- Melis A. Photosystem-II damage and repair cycle in chloroplasts: what modulates the rate of photodamage in vivo?. Trends in plant science. 1999 Apr 1;4(4):130-5.
- Shkrob MA, Yanushevich YG, Chudakov DM, Gurskaya NG, Labas YA, Poponov SY, Mudrik NN, Lukyanov S, Lukyanov KA. Far-red fluorescent proteins evolved from a blue chromoprotein from Actinia equina. Biochemical Journal. 2005 Dec 15;392(3):649-54.
- Langan PS, Close DW, Coates L, Rocha RC, Ghosh K, Kiss C, Waldo G, Freyer J, Kovalevsky A, Bradbury AR. Evolution and characterization of a new reversibly photoswitching chromogenic protein, Dathail. Journal of molecular biology. 2016 May 8;428(9):1776-89.
- Tang Z, Mallikaratchy P, Yang R, Kim Y, Zhu Z, Wang H, Tan W. Aptamer switch probe based on intramolecular displacement. Journal of the American Chemical Society. 2008 Aug 5;130(34):11268-9.
- igem.org [Internet] . Stanford-Brown-Spelman; c2017 [cited 2017 March 5]. Available from: http://2014.igem.org/Team:StanfordBrownSpelman/Cellulose_Cross_Linker.
- Algae UTEXCC. BG-11 Medium [Internet]. UTEX Culture Collection of Algae. Culture Collection of Algae at the University of Texas at Austin; [cited 2017Mar5]. Available from: https://utex.org/products/bg-11-medium
- García‐Borrón JC, Solano F. Molecular Anatomy of Tyrosinase and its Related Proteins: Beyond the Histidine‐Bound Metal Catalytic Center. Pigment Cell Research. 2002 Jun 1;15(3):162-73.
- Chávez-Béjar MI, Balderas-Hernandez VE, Gutiérrez-Alejandre A, Martinez A, Bolívar F, Gosset G. Metabolic engineering of Escherichia coli to optimize melanin synthesis from glucose. Microbial cell factories. 2013 Nov 13;12(1):108.
- com [Internet]. “Lumio Green Detection Kit – Thermo Fisher Scientific.”; c2016 [cited 2016 Oct 2]. Available from: https://www.thermofisher.com/order/catalog/product/LC6090.
- com [Internet]. “FLAG Tag Peptide|Versatile Fusion Tag|CAS# 98849-88-8.”; c2016 [cited 2016 Oct 2]. Available from: http://www.apexbt.com/flag-peptide.html
- Carrard G, Koivula A, Söderlund H, Béguin P. Cellulose-binding domains promote hydrolysis of different sites on crystalline cellulose. Proceedings of the National Academy of Sciences. 2000 Sep 12;97(19):10342-7.
- Nutiu R, Li Y. A DNA–Protein Nanoengine for “On‐Demand” Release and Precise Delivery of Molecules. Angewandte Chemie International Edition. 2005 Aug 26;44(34):5464-7.
- Cornish K, Blakeslee JJ. Rubber Biosynthesis in Plants. American Oil Chemist Society, The Lipid Library. 2011 Nov 2.
- Epping J, van Deenen N, Niephaus E, Stolze A, Fricke J, Huber C, Eisenreich W, Twyman RM, Prüfer D, Gronover CS. A rubber transferase activator is necessary for natural rubber biosynthesis in dandelion. Nature Plants. 2015 Apr 27;1:15048.
- Benedict CR, Madhavan S, Greenblatt GA, Venkatachalam KV, Foster MA. The enzymatic synthesis of rubber polymer in Parthenium argentatum Gray. Plant physiology. 1990 Mar 1;92(3):816-21.
- J Wiemer A, Christine Hsiao CH, F Wiemer D. Isoprenoid metabolism as a therapeutic target in gram-negative pathogens. Current topics in medicinal chemistry. 2010 Dec 1;10(18):1858-71.
- Buhaescu I, Izzedine H. Mevalonate pathway: a review of clinical and therapeutical implications. Clinical biochemistry. 2007 Jun 30;40(9):575-84.
- Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nature biotechnology. 2003 Jul 1;21(7):796.
- Zhao Y, Yang J, Qin B, Li Y, Sun Y, Su S, Xian M. Biosynthesis of isoprene in Escherichia coli via methylerythritol phosphate (MEP) pathway. Applied microbiology and biotechnology. 2011 Jun 1;90(6):1915.
- wikipedia.org [Internet]. Non-mevalonate Pathway; c2016 [cited 2016 June 20]. Available from: https://en.wikipedia.org/wiki/File:Non-mevalonate_pathway.svg
- Nielsen, Alec. Genetic Circuits. BioE 393 Conference; Stanford University.
- Asawatreratanakul K, Zhang YW, Wititsuwannakul D, Wititsuwannakul R, Takahashi S, Rattanapittayaporn A, Koyama T. Molecular cloning, expression and characterization of cDNA encoding cis‐prenyltransferases from Hevea brasiliensis. European Journal of Biochemistry. 2003 Dec 1;270(23):4671-80.
- Black T, Kaskey J. Tire Raw-Material Shortage Triggers Race for Answer [Internet]. Bloomberg.com. Bloomberg; 2014 [cited 2017Mar5]. Available from: https://www.bloomberg.com/news/articles/2014-06-12/tire-raw-material-shortage-triggers-race-for-answer
- Oh SK, Kang H, Shin DH, Yang J, Chow KS, Yeang HY, Wagner B, Breiteneder H, Han KH. Isolation, characterization, and functional analysis of a novel cDNA clone encoding a small rubber particle protein from Hevea brasiliensis. Journal of Biological Chemistry. 1999 Jun 11;274(24):17132-8.
- Xiang Q, Xia K, Dai L, Kang G, Li Y, Nie Z, Duan C, Zeng R. Proteome analysis of the large and the small rubber particles of Hevea brasiliensis using 2D-DIGE. Plant physiology and biochemistry. 2012 Nov 30;60:207-13.
- Rojruthai P, Sakdapipanich JT, Takahashi S, Hyegin L, Noike M, Koyama T, Tanaka Y. In vitro synthesis of high molecular weight rubber by Hevea small rubber particles. Journal of bioscience and bioengineering. 2010 Feb 28;109(2):107-14.
- Liengprayoon S, Bonfils F, Sainte-Beuve J, Sriroth K, Dubreucq E, Vaysse L. Development of a new procedure for lipid extraction from Hevea brasiliensis natural rubber. European journal of lipid science and technology. 2008 Jun 1;110(6):563-9.
- Paul KT. Burning characteristics of materials. Fire and Materials. 1979 Dec 1;3(4):223-31.
- Howell RC, Revskaya E, Pazo V, Nosanchuk JD, Casadevall A, Dadachova E. Phage display library derived peptides that bind to human tumor melanin as potential vehicles for targeted radionuclide therapy of metastatic melanoma. Bioconjugate chemistry. 2007 Oct 2;18(6):1739-48.
- Ballard B, Jiang Z, Soll CE, Revskaya E, Cutler CS, Dadachova E, Francesconi LC. In vitro and in vivo evaluation of melanin-binding decapeptide 4B4 radiolabeled with 177Lu, 166Ho, and 153Sm radiolanthanides for the purpose of targeted radionuclide therapy of melanoma. Cancer Biotherapy and Radiopharmaceuticals. 2011 Oct 1;26(5):547-56.
- Fujishima K, Venter C, Wang K, Ferreira R, Rothschild LJ. An overhang-based DNA block shuffling method for creating a customized random library. Scientific reports. 2015;5.
- Miura Y. Hydrogen production by biophotolysis based on microalgal photosynthesis. Process biochemistry. 1995 Jan 1;30(1):1-7.
- Adams MW. The structure and mechanism of iron-hydrogenases. Biochimica et Biophysica Acta (BBA)-Bioenergetics. 1990 Nov 5;1020(2):115-45.
- Ghirardi ML, Zhang L, Lee JW, Flynn T, Seibert M, Greenbaum E, Melis A. Microalgae: a green source of renewable H 2. Trends in biotechnology. 2000 Dec 1;18(12):506-11.
- Jo JH, Lee DS, Park JM. Modeling and Optimization of Photosynthetic Hydrogen Gas Production by Green Alga Chlamydomonas reinhardtii in Sulfur‐Deprived Circumstance. Biotechnology progress. 2006 Jan 1;22(2):431-7.
- chemstuff.co.uk[Internet].Chemstuff:Tests for gases. (2012). Retrieved 18 May 2016. Available from: https://chemstuff.co.uk/analytical-chemistry/tests-for-gases/
Supporting Information
Supporting information for this work can be found here: Supporting information