iGEM REPORT: Gotta Detect ‘Em All: a multi-STI sensor based on aptamers
Note: This iGEM Report was submitted to the PLOS iGEM Realtime Peer Review Jamboree, and has not undergone formal peer review by any of the PLOS journals. We welcome your comments on this work.
Gotta Detect ‘Em All: a multi-STI sensor based on aptamers
Julien Orlans (1) *, Paul Zanoni (1), Louis Becquey (1), Mathilde Gonthier (1), Mathieu Borel (1), Gianina Ungurean (1), Delphine Bourgeon (1), Margaux Poulalier-Delavelle (1), Jeanne Talonneau (2), Pedro Da Silva (3), Damien Jacques (4), Marie-Pierre Escudié (5), Viviane Chansavang (6), Valérie Desjardin (6), Corinne Dorel (6), Yoann Louis (6) *
- Univ Lyon, INSA-LYON, Département Biosciences, F-69621, VILLEURBANNE, France
- Université Paris-Saclay, ENS-CACHAN, ENSCI Les Ateliers, Telecom Paris-Tech, PARIS, France
- Univ Lyon, INSA-LYON, INRA, BF2I, UMR0203, F-69621, VILLEURBANNE, France
- Univ Lyon, INSA-LYON, Premier Cycle INSA, F-69621, VILLEURBANNE, France
- Univ Lyon, INSA-LYON, Institut Gaston Berger, F-69621, VILLEURBANNE, France
- Univ Lyon, INSA-LYON, Université Lyon1, CNRS UMR5240, MAP, F-69621, VILLEURBANNE, France
*Corresponding author: Julien Orlans (julien.orlans@insa-lyon.fr, julien.orlans@gmail.com), Yoann Louis (yoann.louis@insa-lyon.fr)
Author Contributions
Conceptualization: JO PZ LB MG MB GU DB MPD JT PDS DJ MPE VC VD CD YL
Parts design: MG JO GU CD VC
Aptamers characterization and modifications: MB PZ YL
Streptavidin cloning, production and characterization: MB MG JO GU PZ VC CD
Biomarkers cloning and characterization: MB MG JO PZ VC CD
Revelation systems (fluorescein and latex beads): MB MG JO GU PZ YL CD
Modeling: LB JO PDS
Human practices: PZ DB MPD MPE
Device design: MB MG JT GU VD DJ
Device 3D printing: MB GU PZ DJ
Manuscript writing: LB DB MB CD MG YL JO MPD PZ
Abstract
Nowadays, STIs constitute a major public health issue. Indeed, treatments are often started too late because of belated diagnosis resulting in health problems, such as sterility. If prevention is probably the most effective action one can take to prevent the spread of STIs, early detection could help limit their deleterious effects. In this work, a new diagnosis approach based on aptamers is presented. Bound to paper, they allow the detection of HIV and Hepatitis B biomarkers from a blood sample. The associated device is composed of an anchor, the streptavidin protein, allowing the fixation of the aptamer to the paper via biotin (see graphical abstract). With this system, the HIV-1 Reverse Transcriptase (BBa_K1934060 and BBa_K1934061: protein subunits p51 and p66) and HBsAg (surface antigen of Hepatitis B) are specifically targeted. Then, the biomarker/aptamer complex is detected by two methods. The first one is based on fluorescence. As a proof of concept, a paired ATP/aptamer was used and enabled to successfully detect ATP up to 10 µmol.L-1. However, the signal was not detectable with naked eyes or with a cell phone equipped with blue and green filters either. Therefore, a lateral flow assay with nano-sized latex black beads was tested. This second technique showed that a protein biomarker, such as thrombin, could be complexed with latex beads coated with aptamers, in liquid. Finally, the ultimate step, migration of the latex beads inside paper, needs further optimization. Moreover, to easily handle several STI-tests on a single paper strip, an innovative bio-sourced PLA casing was designed and 3D printed to offer an additional intuitive user-interface.
Financial Disclosure
This work was sponsored by bioMérieux and Fondation INSA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of this manuscript.
Competing Interests
The authors have declared that no competing interests exist.
Data Availability
All data are fully available without restriction: http://2016.igem.org/Team:INSA-Lyon
Introduction
According to the World Health Organization (WHO) key numbers, more than one million Sexually Transmitted Infections (STIs) are acquired every day worldwide [1, 2]. Moreover, the majority of STIs don’t present any symptom that can be easily recognized. Indeed, symptoms of common STIs tend to be non-specific and they typically have a variety of different potential causal agents that may require different treatments. Thus, STIs are a major public health issue and, in order to set up an efficient treatment, rapid diagnosis is essential.
Currently, the STIs are mostly diagnosed thanks to laboratories tests from fluid samples (blood, urine, genital sore, etc) [3]. Diagnostic tests can be divided into at least three different types [1]. First, the most relevant approach to STIs diagnosis is the direct screening of microorganisms themselves (by microscopy, antigen or nucleic acid detection, etc). Secondly, for many important STIs such as HIV and syphilis, screening of antibodies is the favoured diagnostic principle. Thirdly, the last diagnostic tests class detects microbial metabolites, such as molecules altering the pH of specific body fluids. However, these tests commonly allow a relatively late diagnosis, resulting in delayed treatments. If prevention is probably the most effective action one can take to prevent the spread of STIs, early detection can help limit their deleterious effects. As a result, pharmaceutical industries try to develop new detection systems to increase their efficiency and to diagnose people earlier.
The aim of the INSA Lyon 2016 iGEM team project was to elaborate a rapid diagnostic test for multiple diseases and infections. To this end, a paper-based diagnostic test using a drop of blood was developed. This test can be easily handled and used at home without medical supervision, the results should be rapid to obtain and easy to be interpreted. In this study, a diagnosis system for HIV and Hepatitis B is presented. This detection device is based on a new technology: aptamers, which are short single stranded nucleic acids. They basically act as antibodies and present numerous advantages. Compared to antibodies, the process of selection and production of aptamers is considered as quicker, easier, and cheaper [4]. Moreover, they have the advantage to be easily chemically modifiable, so that their affinity for the target can be indefinitely improved. In this work, specific aptamers were designed to target with high specificity the HIV reverse transcriptase [5] and the Hepatitis B surface Antigen (HBsAg) [6]. Two different systems were developed to convert the Aptamer/STI biomarker complexation into a visual output signal: one using fluorescence [7] and the second one using latex beads [8]. The proof of concept of this detection device was made on ATP [9, 10] and thrombin [11-13].
Materials and Methods
Bacterial strain, vector and cultural condition
All plasmids used in the following manipulations were transformed into E. coli NM522. This strain was grown in Luria-Bertani (LB) medium, in SOC (Super Optimal Broth with added glucose) medium or on agar Petri dishes, with the appropriate antibiotics (ampicillin [Amp] = 15 μg.μL-1, kanamycin [Kan] = 15 μg.μL-1 or chloramphenicol [Cm] = 15 μg.μL-1). Used vectors (pSB1C3, pSB1A3, pSB1K3), parts and kit of competent cells were provided by the iGEM Foundation. Restriction enzymes EcoRI, XbaI, SpeI and PstI were provided by the company New England Biolabs (NEB, USA). DNA concentrations were determined using the Nanodrop (ND-1000 Spectrophotometer UV-VIS, No. 5762 series LABTECH).
Gene construction
All parts were designed using SerialCloner software. The three parts of interest were built by assembling different sequences end-to-end: BBa_G00000 prefix/pTAC promotor BBa_K864400 [14], RBS BBa_B0030/coding sequence of interest/double terminator BBa_B0011/suffix BBa_G00001. Endogenous EcoRI, XbaI, SpeI and PstI sites (exclusive of iGEM prefix and suffix) were removed by silent mutation. Moreover, the required spacing between characteristic sequences (for example, between transcription regulated sequences and the TATAA box) were verified. Finally, codons usage was optimized by using the associated application on the Integrated DNA Technologies (IDT, USA) website.The different sequences designed with this method were ordered from IDT as gBlocks Gene Fragment.
Streptavidin with Cellulose Binding Domains (CBDs) part
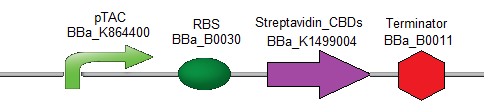
As an anchor for the diagnostic device, the streptavidin [15] protein combined with one or two cellulose binding domain(s) [16] was chosen in order to be easily fixed onto a cellulose paper. The first streptavidin chimeric design contains two Cellulose Binding Domains (CBDs) (Fig. 1).
RFP with Cellulose Binding Domains (CBDs) part
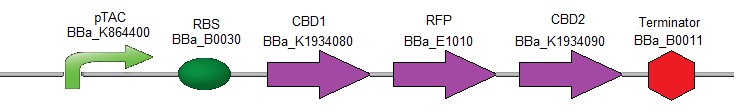
To verify the efficiency of the CBDs for binding to the cellulose, the following composite part was created (Fig. 2). It contains the coding sequences for the two different CBDs (used above) which were fused respectively upstream and downstream of the coding sequence for RFP [17].
Streptavidin with CBD-CipA (Cellulosomal scaffolding protein A)
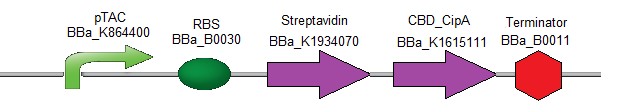
To test another type of CBDs, the following composite part was designed (Fig. 3). It contains the coding sequence of streptavidin followed by the coding sequence of CBD-CipA (Cellulosomal scaffolding protein A) protein. As CBD-CipA [18] shows a better interaction with cellulose, this CBD was preferred to classical CBDs.
Protein purification with CBDs
The Microcrystalline Cellulose (Avicel, Sigma Aldrich) was washed five times in water, and then equilibrated in washing buffer (ammonium sulfate 1 mol.L-1). The culture was pelleted and resuspended in 1 mL of lysis buffer (50 mmol.L-1 Tris, pH = 8, 300 mmol.L-1 NaCl, 10% glycerol). The mix was sonicated on ice, five times for 30 seconds at moderate power. The lysate was centrifuged at 14,000 g for 10 min. The cellulose was packed in small (10 x 10 mm) chromatography columns or syringes barrels. The lysate supernatant was gently poured on the column. Once the liquid started to flow out, regular measurement of the OD280 of the different fractions were performed. Buffer was poured until the OD280 stabilized around zero. The elution was then performed with water. After a short time the OD280 rose, fractions with high OD280 were collected.
Aptamers & quencher
All the aptamers used in these experiments were provided by IDT (USA).
The original sequences of the aptamers used for the Electrophoretic Mobility Shift Assay (EMSA) are :
- Aptamer ATP: 5′- CACTGACCTGGGGGAGTATTGCGGAGGAAGGTTTT -3′ [7]
- Aptamer Thrombin 15nt: 5′- GGTTGGTGTGGTTGG -3′ [11]
- Aptamer Thrombin 29nt: 5′- AGTCCGTGGTAGGGCAGGTTGGGGTGACT -3′ [12]
- Aptamer Thrombin 31nt: 5′- GTGACGTAGGTTGGTGTGGTTGGGGCGTCAC -3′ [13]
- Aptamer HIV Reverse Transcriptase (HIV RT): 5′- TAATACCCCCCCTTCGGTGCTTTGCACCGAAGGGGGGG -3′ [5]
- Aptamer1 HBsAg: 5′- GGGAATTCGAGCTCGGTACCCACAGCGAACAGCGGCGGACATAATAGTGCTTACTACGACCTGCAGGCATGCAAGCTTGG -3′ [6]
- Aptamer2 HBsAg: 5′- GGGAATTCGAGCTCGGTACCCACATGGCATGAAGTATTATTACCCAATTCCATACACAAGCTGCAGGCATGCAAGCTTGG -3′ [6]
- Aptamer3 HBsAg: 5′- GGGAATTCGAGCTCGGTACCGGCACAAGCATATGGACTCCTCTGAACCTACGATGTAGTACCTGCAGGCATGCAAGCTTGG -3′ [6]
For the fixation on paper, a Biotin-TEG function was added on the 3-terminus of each aptamer (3BioTEG) to allow a complex to form between the aptamer and the streptavidin. For the fixation of FITC, an amino modifier C6 function was needed at the 5-terminus (5AmMC6). This function was used on the ATP aptamer. To switch off the light, a quencher was created, it had a reverse complementary sequence to the aptamer 5′-CCCAGGTCAGTG-3′ [7] with a Dabcyl function at the 3-terminus (3Dab). For the fixation of latex deads, the same 5AmMC6 was used on the 15nt and 29nt thrombin aptamers. Their reverse complement for the control band of the test were:
- Reverse complement aptamer thrombin 15nt: 5′-CCAACCACACCAACC-3′
- Reverse complement aptamer thrombin 29nt: 5′-AGTCACCCCAACCTGCCCTACCACGGACT-3′
Electrophoretic Mobility Shift Assay (EMSA)
The aptamer was diluted in folding buffer (SSC 1X) to a final concentration of 200 nmol.L-1. It was denatured at 99 °C, then folded at 42 °C for 20 min and cooled slowly. Serial dilutions of target protein in the binding buffers (see list below) were prepared. 16 µL of proteins were mixed with 4 µL of the folded aptamer. After 20 min at room temperature loading dye was added and 15 µL of the mixture were loaded in each well of a native PAGE gel (12.5% acrylamide, TBE 1X final, 1 mmol.L-1 MgCl, 10 mmol.L-1 KCl). The gel migration was performed at 4 °C at low voltage (100 V maximum). The gel was stained with CYBR Green. Buffer list (pH = 7.4 ):
- Aptamer Thrombin 15 nt: TGKM, Tris 25 mmol.L-1, Gly 192 mmol.L-1,
KCl 0.4 mol.L-1, MgCl2 1 mmol.L-1. - Aptamer Thrombin 29 nt and 31 nt: TGK, Tris 25 mmol.L-1, Gly 192 mmol.L-1,
KCl 0.4 mol.L-1 - Aptamer HbsAg: 20 mmol.L-1 HEPES, 120 mmol.L-1 NaCl, 5 mmol.L-1 KCl,
1 mmol.L-1 CaCl2, 1 mmol.L-1 MgCl2. - Aptamer HIV-RT: 50 mmol.L-1 Tris, 50 mmol.L-1 KCl, 10 mmol.L-1 MgCl2,
1 mmol.L-1 DTT.
Aptamers Modification
FITC on aptamer [19]
1 mmol.L-1 of FITC solution in pure DMSO was prepared. In a microcentrifuge tube: 20 nmol of DNA aptamer modified at the 5 terminus with an amino modifier C6 and 50 µL of 1 mmol.L-1 FITC solution were mixed, and the volume was adjusted to 1 mL with boron (H3BO3 0.1 mmol.L-1, pH = 9) buffer. The mix was incubated overnight at 4 °C.
Fluorescent aptamer purification
The DNA solutions were mixed with 10% v/v LiCl and 250% v/v ethanol (100%) or isopropanol 100% v/v (according to the size of the tube used). The mix was incubated for 10 min prior to centrifugation at 13,000 g for at least 40 min at 4 ºC. The supernatant was removed thoroughly, and 1 mL of ethanol 70% was gently added. After 10 min, the tubes were centrifuged at 13,000 g 10 min, then the supernatant was removed and the pellets air dried.
Quencher on reverse complement aptamer
The purified aptamer and its quencher were mixed in equal amounts in TE buffer, heat shocked at 99 °C for 30 seconds, then hybridized at 42 °C for 30 min.
Latex bead on aptamer [20]
The latex beads (Thermo Fisher, black dyed beads, carboxy modified, 400 nm in diameter) were dissolved in PBS to get a 1 mL, 2% w/v solution. After centrifugation at 15,000 g for 10 min, the beads were resuspended in sulfonic acid buffer (1 mol.L-1, pH 6) by sonication. Very quickly 20 mg of EDC (1-ethyl-3-[3-(dimethylamino)propyl]carbodiimide) was added and the mix was incubated for 15 min. Then a new centrifugation and resuspension in PBS buffer (10 mL) was performed. To 2.5 mL of this solution, 200 µL of a 6 µM DNA solution with a free amine group were added. After a two hour incubation, the mixture was centrifuged and the pellet was washed twice with PBS + Tween 0.01% w/v and BSA 0.1% w/v. Beads can be stored a few weeks in this buffer at 4 °C.
Fluorescence detection
In a liquid phase
To test if the signal of the fluorescent aptamer could be specifically turned off in absence of the ATP target, the aptamer linked to fluorescein, and the quencher fragment were hybridized in SSC 1X at 40 °C for 20 min. 50 µL of a 10 nmol.L-1 aptamer/quencher solution was prepared this way. A serial dilution of ATP in PBS pH 7.1 was prepared (ranging from 10 mmol.L-1 to 1 µmol.L-1). 50 µL of each ATP dilutions were poured in each well, in triplicates. Controls were: plain PBS and a 10 mmol.L-1 GTP solution. The hybrid DNA was diluted in 1 mL of PBS, and quickly added (50 µL) to each well with ATP or control solutions. Fluorescence was measured (490 nm excitation, 530 nm emission) with a plate reader within 5 min to prevent bleaching.
On a paper substrate
The FITC labelled DNA was prepared as described, it was resuspended in 100 µL of TE buffer, 25% v/v loading dye was added, and the mix was distributed in nine wells of a native 15% acrylamide PAGE. 330 ng of DNA was used per well. A Southern Blot experiment was then carried out following standard procedure [21]. During the hybridization phase, 5 µg of the quencher DNA fragment was used. Once the membrane was hybridized and washed, 10 µL of ATP solution (in different concentrations) were poured very carefully on every lane. The solutions ranged from 100 mmol.L-1 to 1 µmol.L-1. After five minutes, the membrane was washed twice with PBS, and the fluorescence imaged with a ChemiDoc system (BioRad).
Latex beads experiments
To test the beads, 96-well plates covered with streptavidin were used. 100 µL of a PBS solution containing 1 ng of DNA were poured in each well. DNAs to be tested include the reverse complements of thrombin aptamers, and the two tested aptamers, they are biotynilated. After
1 h at 30 °C the wells were washed three times with binding buffer (PBS buffer + 0,01 % Tween + 0.1 % BSA + 1 mmol.L-1 MgCl2). In each well, 20 µL of the latex beads solution were poured (the beads were coated with the complementary aptamer of the corresponding ones in the wells), and 70 or 80 µL of binding buffer. In the test wells, 10 µL of thrombin were added (several concentrations are tested). After 30 min on a rotating plate at room temperature, the wells were washed with PBS, very carefully. A measure of the OD600 of each well was done.
Results
Designing an anchoring system on paper
To avoid the diffusion of the aptamer sensor on paper, a protein complex is needed to anchor the aptamer on the paper surface. Chimeric proteins containing C-terminus cellulose binding domain (CBD) with a cellulosome scaffolding protein A, called CipA, were engineered. To select the best CBD candidate, ability to bind cellulose of the streptavidin-CBD (BBa_K1934020, INSA Lyon 2016 iGEM team) and of the streptavidin-CiPA (BBa_K1934030, INSA Lyon 2016 iGEM team) was compared (Fig. 4).
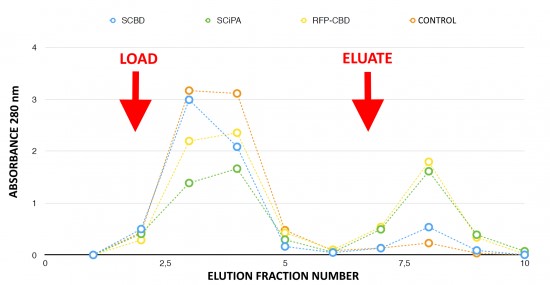
Fraction 8 corresponding to the elution peak contained the majority of the purified proteins. This fraction was collected for further testing. The peak corresponding to Streptavidin-CipA (green marks) in fraction 8 is higher than the Streptavidin-CBD peak (blue marks). Chimeric proteins stick to the cellulose until elution, meaning that more Streptavidin-CipA has been retained in the column during the washing run. So, Streptavidin-CipA has better adsorption properties with cellulose than Stanford’s existing part. The RFP-CBD acts as a positive control with visual follow-up thanks to the red color given by the fixation of the protein to the column. No retention could be observed in the control experiment in absence of Cellulose-Binding-Domains.
Functionalization of the aptamers
In order to control the interaction of the aptamers with other pieces of the detection system, functionalization was achieved by grafting additional components to aptamers. The ATP-aptamer was decorated with fluorescein thanks to crosslinking in presence of FITC. To temporarily switch off the fluorescence, a non fully-affine reverse-complement quencher molecule (DABCYL) was linked. DABCYL quenches the fluorescence by Fluorescent Resonance Energetic Transfert. When present, ATP displays higher affinity for the aptamer and is therefore able to displace the quencher, resulting in a fluorescent signal.
Such a labelling allows further detection of the aptamer/target complex. The aptamer used here is known as ATP aptamer [7]. The mix was analyzed on a PAGE gel.
Figure 5 shows the overlays of ethidium bromide staining of the PAGE gel (red) and fluorescence imaging (purple). From the two UV visible bands after ethidium bromide staining, only the heaviest band was fluorescent. This result indicates that the fluorescein was successfully crosslinked to the aptamers. A significant amount of untagged aptamers was still observed, showing that the reaction is not complete.
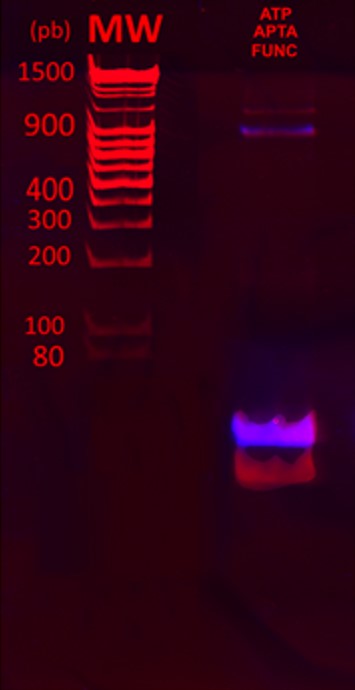
Binding functionalized aptamers to protein complex
The fixation of functionalized aptamers onto a support is enabled by a Biotin/Streptavidin link. The biotin non-covalent link between a fluorescent DNA oligo (FITC on the 5′ end and biotin on the 3′ end) and 3 different anchoring complexes was compared (Fig. 6).
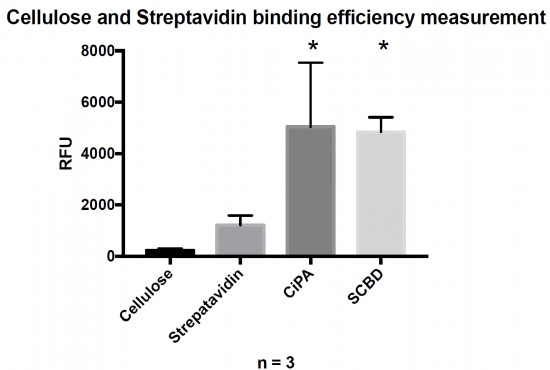
Fluorescence of the reporter system is proportional to the amount of bound protein. Binding cellulose with the engineered proteins was statistically different than raw streptavidin (Student’s test with p-value < 0.05).
The cellulose alone showed no fluorescence, thus confirming that aptamers cannot be directly linked to paper. A protein anchoring system is required. The streptavidin experiment shows a spontaneous ability of streptavidin to link a small fraction of cellulose. Both engineered streptavidin-CBDs allowed the formation of complexes with the fluorescent reporter system and cellulose that could be detected by measuring the green fluorescence. There was no significant difference in terms of binding performance between the two engineered streptavidin designs. Nevertheless, the binding capacity of both protein was significantly higher than streptavidin alone demonstrating the efficiency of the system.
Electrophoretic Mobility Shift Assays
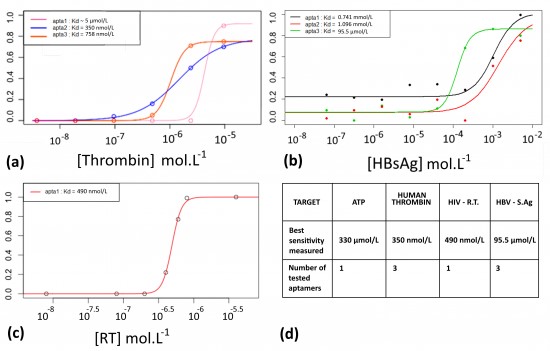
To both demonstrate the ability of the aptamers to fix their target, and measure their affinity constants, Electrophoretic Migration Shift Assays (EMSA) were carried out. A proof of concept was realized with a well described aptamer and its target protein: the human thrombin. This experiment allows to measure the affinity of an aptamer with its target. The aptamer/protein complex formed in presence of increasing amount of proteins was separated and quantified on native PAGE. Figure 7(a) shows the EMSA results for three different aptamers with affinity for the thrombin. The curves were modeled to fit the points (thanks to a 4 parameters model and the nls fonction of R). This allowed to calculate the Kds, which are concentration values corresponding to the inflexion points. The most sensitive thrombin aptamer, aptamer of 31 nt, detected thrombin down to 350 nmol.L-1. Based on these results, EMSAs were performed with two STI-biomarkers/aptamers couples (HIV-1 Reverse-Transcriptase and HBsAg), see Figures 7 (b), (c) and the best sensitivity associated with each biomarker is reported in the table of the Figure 7 (d).
Proof of concept of the detection with FITC
Fluorescent labelled ATP aptamers (with FITC) were loaded and separated on a PAGE gel. A constant amount was loaded in each well. Figure 8 shows that the fluorescent response to ATP concentration is semi-quantitative and follows a sigmoid curve (in liquid samples).
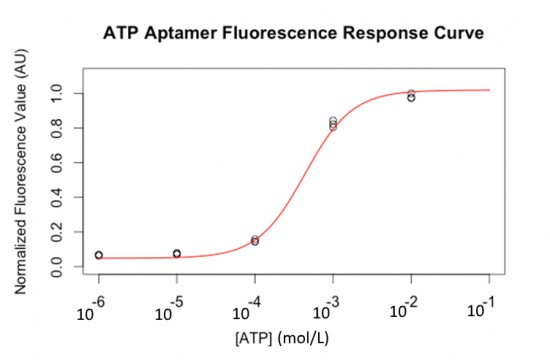
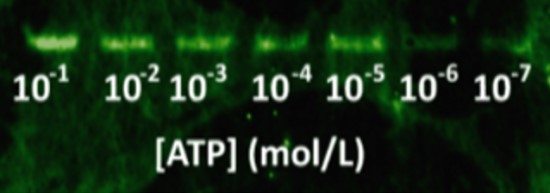
Then, DNA was blotted on nitrocellulose using capillary transfer, with quencher oligos to block fluorescence (a reverse complement fragment of 6 nucleotides of the aptamer labelled with a DABCYL molecule at its 3’ end). This quencher oligo was hybridized in situ by incubating the membrane in hybridization buffer, similar to the buffer for Southern Blots [21]. After washing the quencher excess, increasing amounts of ATP gradient were applied on each lane of the nitrocellulose membrane. The blotting of the complete system on a nitrocellulose sheet with a range of ATP concentrations is shown in Figure 9. The ATP aptamer successfully detects ATP down to 10 µmol.L-1.
Proof of concept with a second detection system that uses latex beads
In this second version of revelation system, the target molecule is taken in sandwich between two aptamers that link two different apitopes. The first aptamer is fixed on black dyed nano-sized latex beads. The second one is fixed on the support and catches the target when the assay flow comes by capillarity (Fig. 10). With this strategy, the user does not need any fluorescence measurement to detect aptamers: as the latex beads are black, they are visible with naked-eye.
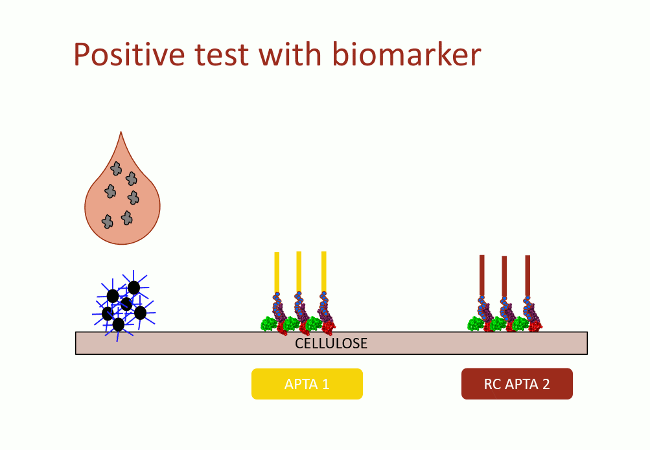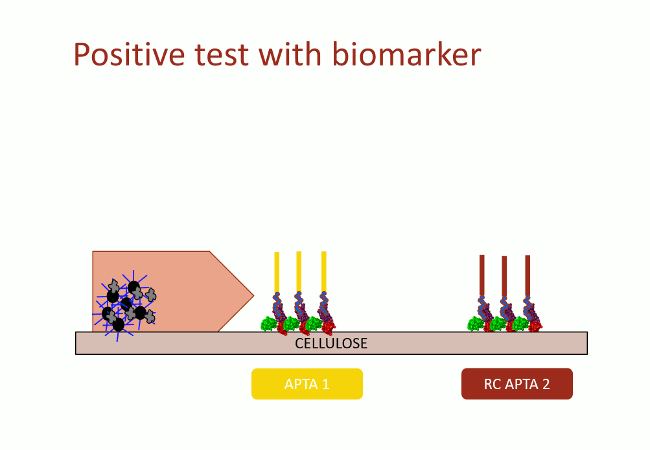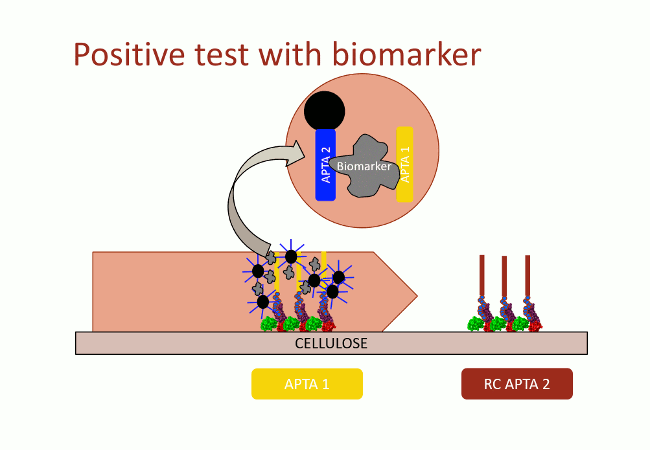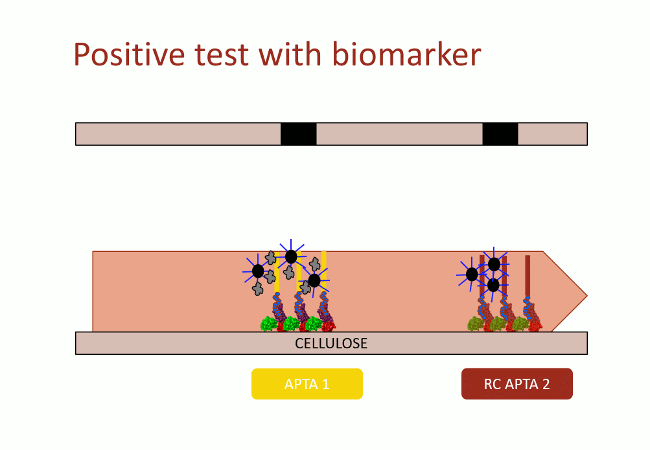
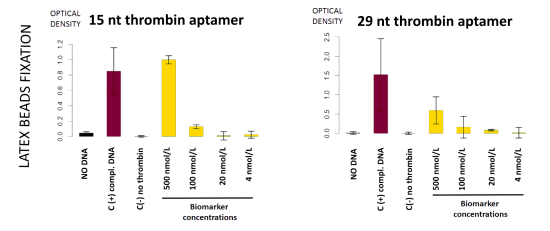
This detection system was firstly implemented on streptavidin-coated plates. The beads fixation to the streptavidin-coated well bottom results in darkening the well. This phenomenon was quantified by OD600 measurement. On Figure 11, in absence of DNA, beads did not stick
(bar 1 in black). Beads with complementary strands aptamers allowed a good fixation, and constituted the positive control (bar 2 in red). The full detection system in absence of biomarker (thrombin), the negative control, did not allow fixation of the beads (bar 3 in grey). Decreasing amount of thrombin allowed to determine the sensibility of the test, i.e. 100 nmol.L-1 in these conditions (yellow bars).
Discussion
Developing a lateral flow test requires four modules: i) A biomarker referred to as target; due to experimental or safety issues, a well described molecule was used in place of the STI-biomarker to elaborate the proof of concept. ii) A paired aptamer to catch the biomarker. iii) An anchor to fix the aptamer-biomarker complex to the support. iv) A visual output signal. Our approach consists in setting and characterizing each module before assembling the whole system together.
We were able to assemble a functional system using ATP as target and a functionalized aptamer (module ii)) doubled tagged with biotin (module iii)) and fluorescein (module iv)) (Fig. 5). The chimeric streptavidin and the aptamer put together do not lose their individual properties, showing the pertinence of our modular approach. Our results showed that the system is able to successfully detect ATP with a better affinity constant in liquid (330 µmol.L-1) than on nitrocellulose support (10 µmol.L-1). The sensibility is actually lowered by a 30 factor on cellulose, probably due to the diffusion of the target molecules on the paper. These results could be improved by finding a compromise between the affinity of the ATP and the reverse complement to aptamer in order to ensure a specific recognition with a low detection threshold. Moreover, paper substrate was shown to emit auto-fluorescence (Fig. 9). This fluorescent noise competes with the emitted signal and lower the detection capacities of the system. Screening of cellulosic substrate with low fluorescence could also improve the system sensitivity.
To develop a more sensitive system and a proof of concept with a proteic biomarker more representative of the chosen STI biomarkers (HIV-RT and HbsAg), a system using Thrombin as a target (module i)) and latex beads (module iv) was successfully assembled and successfully assayed in liquid in streptavidin coated plates (Fig. 11). HIV-RT, HBsAg and thrombin have in common to be recognized by 2 different and specific aptamers. This property allows these 3 biomarkers to form sandwiches with aptamers carrying different functions and is the basis of our detection system. Preliminary results obtained with HIV-RT and HBsAg indicate that module i) and ii) are functional. The complementary modules iii) and iv) have now to be added and the whole system assayed. Compared to fluorescent output, the revelation system involving black latex beads ensures an optimal reading condition for blood samples [22] and is not affected by the paper self-fluorescence.
Regarding Thrombin, we have shown that the Kd can go down to 7.2 nmol.L-1 [13]. The procedure can be improved by enhancing the affinity of the aptamer, i.e. by introducing mutation in the DNA sequence or using a SELEX method, or by optimizing the ionic concentration of the folding buffer [4]. Improvement of the antibody-antigen complex Kd could reach 2.4 nmol.L-1 [23]. Such an increase of the detection threshold could allow an earlier detection of the biomarker. This issue is crucial to cure and prevent the sexually transmitted infections development.
The Gotta Detect‘ Em All test was released with the conscience of the issues related to the false positive and false negative risk. Even if the principle of the test is quite complicated, it is dedicated to limit the undesired interaction, which is crucial to increase the reliability of the detecting system.
The main issue is that our detection procedure requires two different aptamers, and the sensitivity of the method will be limited by the highest Kd (less affinity) between the two aptamers. The need of two aptamers can be seen as a limitation of the procedure. Indeed only one aptamer is often described for one biomarker.
Conclusion
Aptamers are a flexible tool, with a promising future. Utilizing this technology in a device aimed to be broadcasted vastly is nevertheless complex. The interface between aptamers interactions with their targets and the end user is difficult to set up. Here we have shown that two techniques could be implemented to solve this problem. Fluorescence detection is working, but with a poor yield and the latex beads lateral flow system is promising. These both systems would need further improvements to be implemented in detection devices commercially available. This technology is nevertheless promising, offering good results at a low price.
Acknowledgments
First we would like to thanks all the participants of this project who help us during the summer 2016: Mickaël Hubert, Thomas Sabate and Marianne Chouteau.
We also want to thank the following people for their kind help with our project. Their support was precious to us and they gave us a lot of good advice! So a huge thanks to:
Dr. Philippe Oger for his great expertise about bacterial behaviour and protein production, Prof. Rainer Bischoff for his guidance about biomarkers, Dr. Anne Ebel for her guidance about understanding how to calculate the viral load for HIV and HBV, Dr. Philippe Lejeune for his numerous advice concerning bacterial cultures and handling, Dr. Sebastien Lemaire for his knowledge about yeasts, Dr. Ciara O’Sullivan for all her knowledge about aptamers and paper-based tests, Dr. Florence Popowycz for her clarification about organic chemistry with complex molecules such as DNAs, Dr. Sylvie Reverchon for her guidance about the SELEX method and its utility for choosing aptamers, Dr. Christophe Soulage for his kind help with getting blood, Dr. Anna Zaidman and Mrs Karen Gaget for the Flow Cytometer training, Mr. Davyd Chaumard for his help in the preparation of the presentation of the project, Mr. Cédric Grossi for the rabbit blood he provided for our tests, Mr. Sebastien Orlans for his support with light filters, Mrs Samira Trouilleux for her experience and her valuable proposals, Mrs Céline Viollet, Amandine Barbier, Caroline Charre and Pr. Christian Chidiac and the whole team of the CeGiDD Croix-Rousse in Lyon for their precious analyses and for their kindness.
Special thanks to our sponsors: bioMerieux and Fondation INSA.
Response to Reviewers
A transcript of the reviewer comments and author responses from the Live Peer Review Jamboree can be found here: INSA-Lyon Response to Reviewers
References
- Sexually transmitted infections (STIs) [Internet]. World Health Organization. 2017 [cited 26 January 2017]. Available from: http://www.who.int/mediacentre/factsheets/fs110/en/
- Long L, Abraham C, Paquette R, Shahmanesh M, Llewellyn C, Townsend A and Brief interventions to prevent sexually transmitted infections suitable for in-service use: A systematic review. Preventive Medicine. 2016;91:364-382.
- Yetisen A, Akram M, Lowe C. Paper-based microfluidic point-of-care diagnostic devices. Lab on a Chip. 2013;13(12):2210.
- Darmostuk M, Rimpelova S, Gbelcova H, Ruml T. Current approaches in SELEX: An update to aptamer selection technology. Biotechnology Advances. 2015;33(6):1141-1161.
- Miller M, Tuske S, Das K, DeStefano J, Arnold E. Structure of HIV-1 reverse transcriptase bound to a novel 38-mer hairpin template-primer DNA aptamer. Protein Science. 2015;25(1):46-55.
- Xi Z, Huang R, Li Z, He N, Wang T, Su E and Selection of HBsAg-Specific DNA Aptamers Based on Carboxylated Magnetic Nanoparticles and Their Application in the Rapid and Simple Detection of Hepatitis B Virus Infection. ACS Applied Materials & Interfaces. 2015;7(21):11215-11223.
- Su S, Nutiu R, Filipe C, Li Y, Pelton R. Adsorption and Covalent Coupling of ATP-Binding DNA Aptamers onto Cellulose. 2007;23(3):1300-1302.
- Jauset-Rubio M, Svobodová M, Mairal T, McNeil C, Keegan N, El-Shahawi M and Aptamer Lateral Flow Assays for Ultrasensitive Detection of β-Conglutin Combining Recombinase Polymerase Amplification and Tailed Primers. Analytical Chemistry. 2016;88(21):10701-10709.
- Huizenga D, Szostak J. A DNA Aptamer That Binds Adenosine and ATP. 1995;34(2):656-665.
- Lin C, Patei D. Structural basis of DNA folding and recognition in an AMP-DNA aptamer complex: distinct architectures but common recognition motifs for DNA and RNA aptamers complexed to AMP. Chemistry & Biology. 1997;4(11):817-832.
- Padmanabhan K, Padmanabhan K P, Ferrara J D, Sadler J E, Tulinsky A. The structure of alpha-thrombin inhibited by a 15-mer single-stranded DNA aptamer. The Journal of Biological Chemistry. 1993;268:17651-17654.
- Russo Krauss I, Pica A, Merlino A, Mazzarella L, Sica F. Duplex–quadruplex motifs in a peculiar structural organization cooperatively contribute to thrombin binding of a DNA aptamer. Acta Crystallographica Section D Biological Crystallography. 2013;69(12):2403-2411.
- Russo Krauss I, Spiridonova V, Pica A, Napolitano V, Sica F. Different duplex/quadruplex junctions determine the properties of anti-thrombin aptamers with mixed folding. Nucleic Acids Research. 2015;44(2):983-991.
- De Boer H, Comstock L, Vasser M. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proceedings of the National Academy of Sciences. 1983;80(1):21-25.
- Fairhead M, Krndija D, Lowe E, Howarth M. Plug-and-Play Pairing via Defined Divalent Streptavidins. Journal of Molecular Biology. 2014;426(1):199-214.
- Linder MTeeri T. The cellulose-binding domain of the major cellobiohydrolase of Trichoderma reesei exhibits true reversibility and a high exchange rate on crystalline cellulose. Proceedings of the National Academy of Sciences. 1996;93(22):12251-12255.
- Shu X, Shaner N, Yarbrough C, Tsien R, Remington S. Novel Chromophores and Buried Charges Control Color in mFruits. Biochemistry. 2006;45(32):9639-9647.
- Yaniv O, Morag E, Borovok I, Bayer E, Lamed R, Frolow F and Structure of a family 3a carbohydrate-binding module from the cellulosomal scaffoldin CipA ofClostridium thermocellumwith flanking linkers: implications for cellulosome structure. Acta Crystallographica Section F Structural Biology and Crystallization Communications. 2013;69(7):733-737.
- Introduction to Amine Modification – Section 1.1 | Thermo Fisher Scientific [Internet]. Thermofisher.com. 2017 [cited 21 January 2017]. Available from: https://www.thermofisher.com/fr/fr/home/references/molecular-probes-the-handbook/fluorophores-and-their-amine-reactive-derivatives/introduction-to-amine-modification.html#
- Hermanson G. Bioconjugate techniques. 1st Amsterdam: Academic Press; 2008.
- Southern E. Southern blotting. Nature Protocols. 2006;1(2):518-525.
- Bangs L. New developments in particle-based immunoassays: Introduction. Pure and Applied Chemistry. 1996;68(10).
- Hrkal Z, Cajthamlová H, Novák J, Paluska E, Stöckbauer P. Monoclonal Antibodies Against Human Antithrombin III. Hybridoma. 1991;10(5):633-640.